One of the important enzymes in the replication cycle of the SARS-CoV-2 virus is the helicase, which is also known as non-structural protein 13 (nsp13). During the viral life cycle, the holo-RNA-dependent RNA polymerase, also known as nsp12, is thought to coordinate with several additional factors, including the nsp13 helicase (Snijder et al., 2016; Sola et al., 2015) as shown in figure 1.
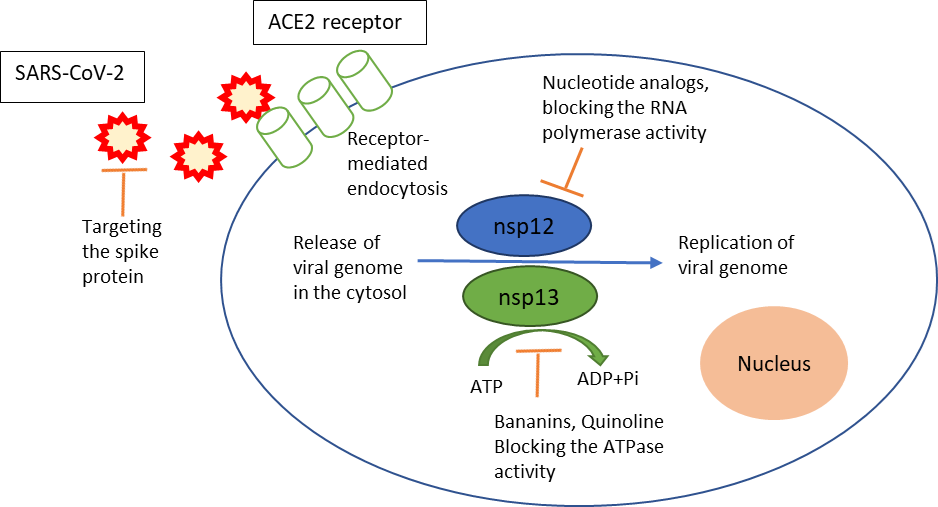
Figure 1. The possible targets for drug development against SARS-CoV-2. Adapted from Habtemariam et al., 2020.
The ATPase domain of nsp13 catalyzes an NTP-dependent unwinding of duplex RNAs into single strands in the 5’ to 3’ direction. (Habtemariam et al., 2020, Snijder et al., 2016) Previous reports on potent inhibitors targeting the ATPase domain of herpes simplex virus helicase suggest that helicase enzyme is chemically tractable. (Kleymann et al., 2002)
This post provides an overview of the druggability and amino acid variability of both the helicase ATP binding site and its central channel where the RNA molecule binds (figure 2) using the SARS-CoV-2 nsp13 helicase structures available from the protein data bank (PDB).
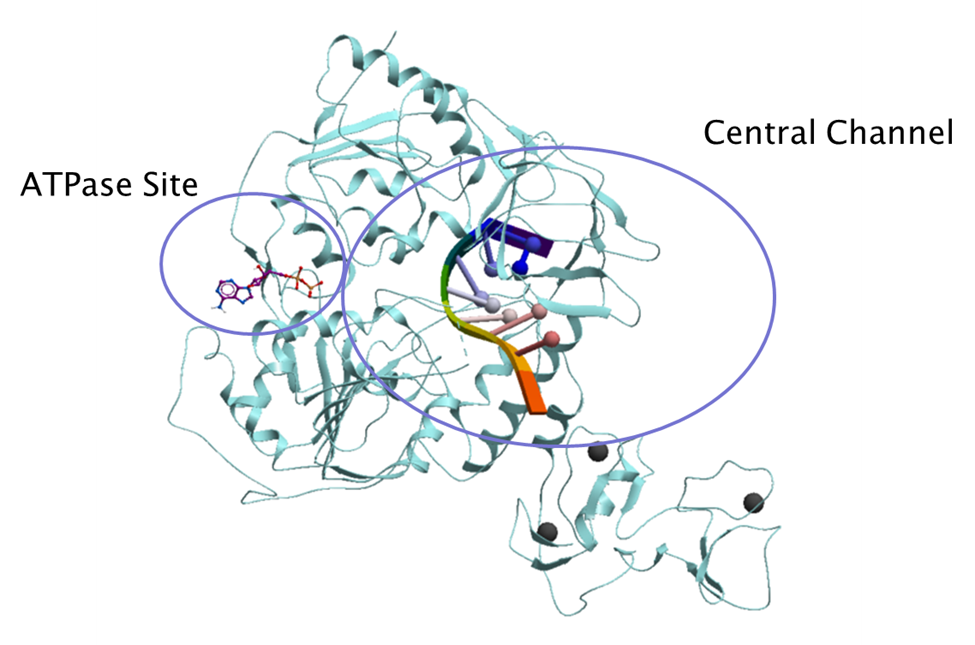
Figure 2. Nsp13 helicase ATPase site and central channel (PDB: 6zsl). The RNA molecule is shown in ribbon diagram from the superimposed Upf1 complex (PDB code: 2xzl) and the ADP molecule shown in purple stick representation superimposed from SARS-CoV-2 nsp13 helicase structure bound to ADP (PDB: 6xez).
Using SiteMap (Schrodinger, NY) we assessed the druggability of the binding sites using the druggability score (Dscore). The ATP binding site of nsp13 and central channel have Dscores of 1.054 and 1.091 respectively. Both Dscores are indicative of druggable sites.
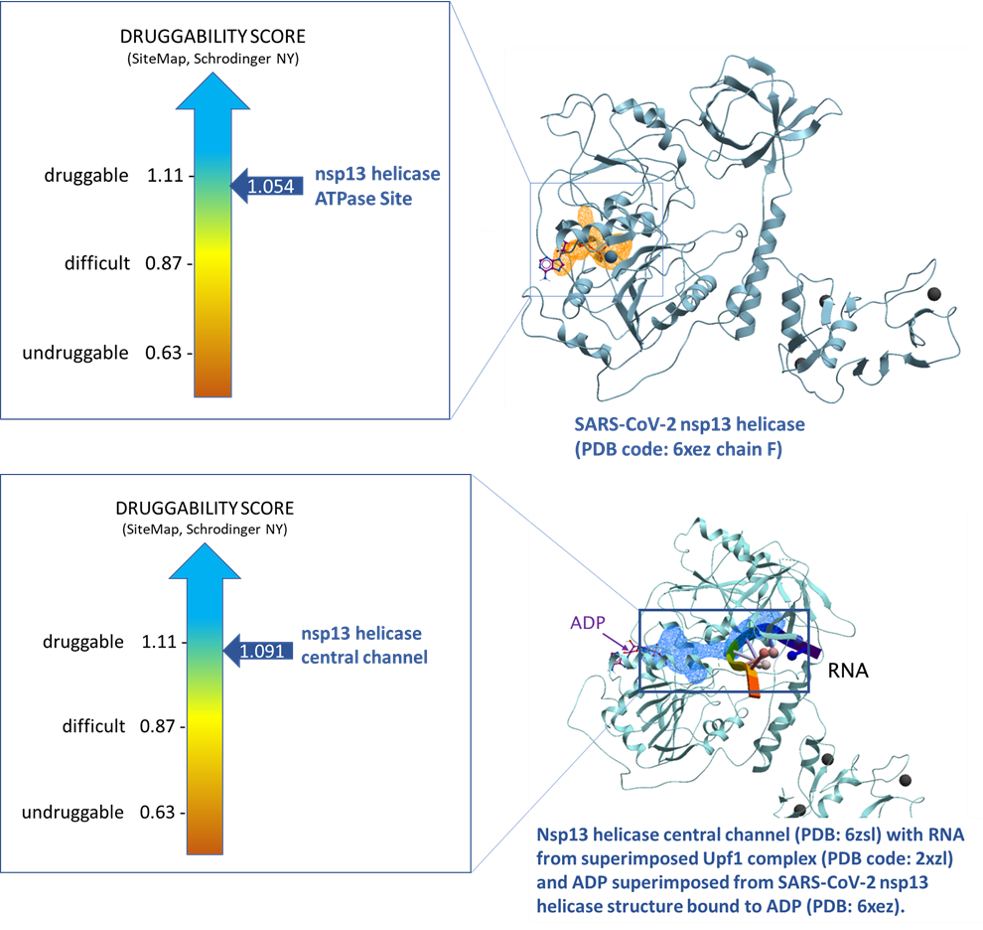
Figure 3. Druggability analysis of SARS-CoV-2 nsp13 helicase ATPase site (top panel, PDB: 6xez chain F) and central channel (bottom pane, PDB: 6zsl).
We then determined the residues that line the nsp13 helicase ATPase site and central channel using ICM PocketFinder (Molsoft, San Diego) and the nsp13 helicase structures. We identified 38 amino acid sidechains lining the nsp13 ATPase site (figure 5, Zenodo report) and 25 amino acid sidechains lining the central channel (figure 6, Zenodo report).
We conducted a broad survey of viral proteins from Alpha- and Betacoronaviruses to identify the most conserved druggable sites. By integrating variability and druggability data, we can then infer the most promising strategies for developing broad-spectrum viral inhibitors. This analysis can be valuable in the context of the emergence of future coronaviruses from circulating viral strains in bat reservoir species. We looked at twenty-seven reviewed nsp13 sequences from Uniprot, where six belong to Alphacoronavirus genus entries, and twenty-one belong to the Betacoronavirus genus. We then assessed the variability of amino acid lining the nsp13 helicase ATP binding site and its central channel by performing multiple sequence alignment.
We made diversity dendrograms focusing on the 38 sidechains of the ATPase site (figure 7, zenodo report) and the 25 sidechains of the central channel (figure 8, zenodo report) from step2. At the ATPase site, 26 out of 38 residues were found to be identical across these 27 entries which translates to 68% identity, indicating a relatively conserved site. At the nsp13 central channel, only 3 out of 25 residues were non-identical; 88% of residues were identical. This shows that the nsp13 helicase central channel where the RNA molecule binds is highly conserved.
We assessed the genetic variability of the ATPase site and the central channel of nsp13 among SARS-CoV-2 samples. Nicola De Maio, our collaborator from Nick Goldman’s lab at the European Bioinformatics Institute, looked at more than 39000 SARS-CoV-2 genome samples from COVID-19 patients and identified all mutations at the two sites of interest of nsp13 helicase. He identified 12 non-synonymous variants at the ATPase site (table 1, zenodo report) and no non-synonymous variant at the central channel of nsp13 helicase.
We conclude that the ATPase site of SARS-CoV-2 nsp13 helicase is less conserved than its RNA-binding central channel across Alpha- and Betacoronaviruses. We observe a similar pattern by looking at non-synonymous variants across SARS-CoV-2 samples from COVID-19 patients. While the ATPase site where ADP binds is druggable and relatively conserved (%68), the central channel of nsp13 helicase where RNA binds is druggable and highly conserved (88%). Our finding supports the design of pan-coronavirus inhibitors targeting the central channel of nsp13 helicase.
You can view the full report on Zenodo.
Please contact me via the “Leave a comment” link at the top of this post. Stay Tuned for more updates on this project.
References
Habtemariam, S., Nabavi, S. F., Banach, M., Berindan-Neagoe, I., Sarkar, K., Sil, P. C., & Nabavi, S. M. (2020). Should we try SARS-Cov-2 Helicase inhibitors for COVID-19 therapy? Archives of Medical Research, 51(7), 733-735. https://doi.org/10.1016/j.arcmed.2020.05.024
Kleymann, G., Fischer, R., Betz, U. A., Hendrix, M., Bender, W., Schneider, U., Handke, G., Eckenberg, P., Hewlett, G., Pevzner, V., Baumeister, J., Weber, O., Henninger, K., Keldenich, J., Jensen, A., Kolb, J., Bach, U., Popp, A., Mäben, J., … Rübsamen-Waigmann, H. (2002). New helicase-primase inhibitors as drug candidates for the treatment of herpes simplex disease. Nature Medicine, 8(4), 392-398. https://doi.org/10.1038/nm0402-392
Snijder, E., Decroly, E., & Ziebuhr, J. (2016). The Nonstructural proteins directing coronavirus RNA synthesis and processing. Coronaviruses, 59-126. https://doi.org/10.1016/bs.aivir.2016.08.008
Sola, I., Almazán, F., Zúñiga, S., & Enjuanes, L. (2015). Continuous and discontinuous RNA synthesis in coronaviruses. Annual Review of Virology, 2(1), 265-288. https://doi.org/10.1146/annurev-virology-100114-055218

Nice work! I look forward to discussing further 🙂
Thanks! I look forward to sharing more of our work with you!