In two of my previous posts 13 and 14, I showed how we found the residues lining the ADP ribose (ADPr) binding site of SARS-CoV-2 nsp3-Mac1 and we looked at the variability of this site across Alpha- and Betacoronavirus entries from UniProt.
In addition to the UniPro sequences, we were interested in looking at variants from COVID-19 patients. Nicola De Maio, our collaborator from Nick Goldman’s lab at EBI, looked at more than 15000 SARS-CoV-2 samples and identified all mutations at the ADPr binding site.
He identified 15 non-synonymous variants as shown in table 1. For example, at residue number 23, the wild-type sidechain in SARS-CoV-2 Mac1 is isoleucine. In the sample batch Nicola has looked at, he saw that 15872 samples had isoleucine at that position while there were 2 samples with other variants at this position; one was a valine and another was a serine.
| Number | Wild Type | Non-synonymous Variants |
| 1 | Ile23 | I (15872), V (1) |
| 2 | Ile23 | I (15872), S (1) |
| 3 | Ala39 | A (15872), V (3) |
| 4 | Asn40 | S (1), N (15874) |
| 5 | His45 | Y (1), H (15874) |
| 6 | Gly46 | *(1), G (15874) |
| 7 | Gly48 | C (1), G (15873) |
| 8 | Val49 | I (2), V (15871) |
| 9 | Ala52 | A (15873), V (1) |
| 10 | Ala129 | A (15781), V (1) |
| 11 | Ile131 | I (15781), F (1) |
| 12 | Phe132 | V (1), F (15781) |
| 13 | Pro136 | P (15760), L (6) |
| 14 | Pro136 | P (15760), S (6) |
| 15 | Leu160 | L (15870), F (1) |
Table 1. The non-synonymous variants at Mac1 ADPr binding site across more than 15000 SARS-CoV-2 samples. * denotes that the mutation has caused a premature stop codon at position Gly46, and the patient’s status with this mutation in their viral sample was marked as “released”.
The next step for us was to perform energy calculations for the SARS-CoV-2 variants at nsp3-MAC1 ADPr binding site in order to predict their effects on ADPr binding.
To predict the effect of these mutations on the binding of a known Mac1 ligand, ADPr (PDB: 6w02), we estimated the difference in the binding energy of the ligand between the wild type and the mutated forms of the protein with ICM (Molsoft, San Diego). ICM calculates the change in Gibbs binding energy (dGbind in kcal/mol) of the ligand as the energy of the protein-ligand complex minus the energy of the isolated protein and ligand. The difference in binding energy (ddGbind), which is what we are interested in, is dGbind [mutant] minus dGbind [wild-type]. Since the precision of the method is about 2 kcal/mol, ddGbind values > 2 kcal/mol indicate mutations that significantly penalize ligand binding. (Schapira et al., 1999) The color-coded structure in figure 1 highlights the variants among the SARS-CoV-2 samples at ADPr binding site of nsp3-Mac1 based on their effects on ligand binding.
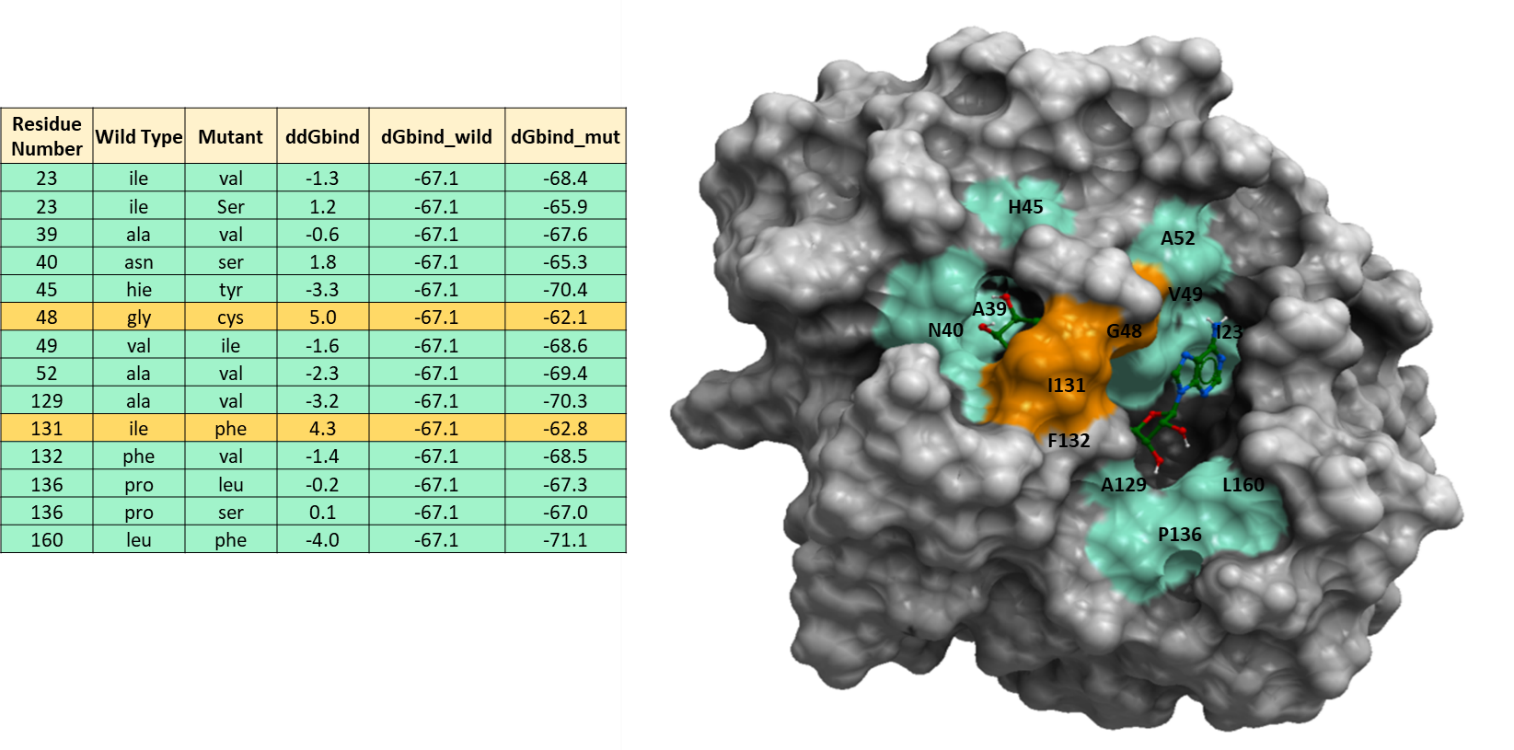
Figure 1. Energy calculations of non-synonymous mutations at SARS-CoV-2 nsp3-Mac1 ADPr binding site to predict their effect on ADPr binding. The energy unit for ddGbind values is Kcal/mol. The green color represents residues with mutations with ddGbind values below 2.0 kcal/mol and orange color shows residues with mutations that have ddGbind values greater than 2.0 kcal/mol.
Looking at figure 1, we see that the ddGbind values for G48C, I131F are above 2.0 Kcal/mol which is considered a significant change in binding according to a previous study. (Schapira et al., 1999) The remainder of the variants are predicted to have minor effects on ligand binding (-2kcal/mol < ddGbind < 2kcal/mol).
Based on previous studies, coronaviruses with mutations in their macrodomains are highly attenuated and in vivo cause minimal disease. This is due to the role of macrodomain in reversing ADP-ribosylation which disrupts the host’s immune response. (Grunewald et al., 2019)
Below, I use surface representation to model the two SARS-CoV-2 nsp3-Mac1 variants, G48C (figure 2) and I131F (figure 3), as visualization aids. In my Zenodo report, you can also view the modeled mutations using sticks representation.
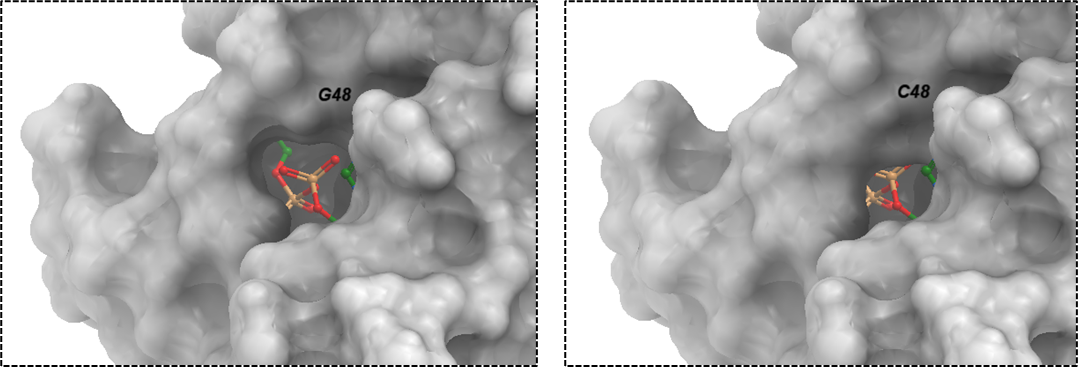
Figure 2. The surface representation of SARS-CoV-2 macrodomain variant: G48C. The corresponding ddGbind value for this mutation is 5.0 Kcal/mol.
C48 is a bulkier residue than glycine as shown in figure 2 which could be the reason for unfavorable and destabilizing predicted effect on ADPr binding with the nsp3-Mac1 at ddGbind value of 5.0 Kcal/mol. The diphosphate moiety binds to the glycine-rich segment (Gly46-Gly47-Gly48) loop on the protein which provides stabilizing effect for the proximal ribose ring in the pocket through hydrogen bonds with G46 (OH2’), G48 (OH1’). (Michalska et al., 2020)
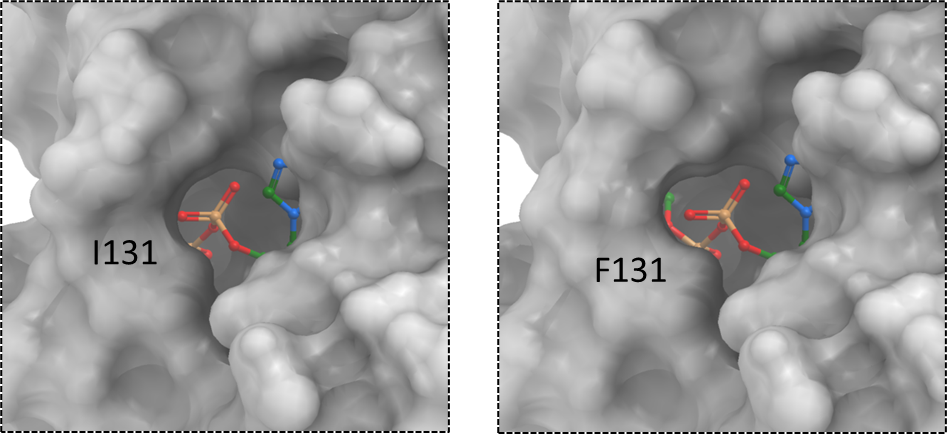
Figure 3. The surface representation of SARS-CoV-2 macrodomain variant: I131F. The corresponding ddGbind value for this mutation is 4.3 Kcal/mol.
Although phenylalanine is a bulkier residue than isoleucine, its bulkiness is protruded outside the pocket rather than inside the pocket where the phosphate linker is located. According to Karolina Michalska and their colleagues, the proximal ribose ring is stabilized in the pocket by hydrophobic interactions with I131. The mutation to phenylalanine may disturb this hydrophobic interaction. (Michalska et al., 2020)
To view the 2D ligand-protein diagram showing the chemical interactions between ADPr and nsp3-Mac1, please refer to figure 11 on my Zenodo report.
We see a lot of structural diversity at the ADP-ribose binding site of the first macrodomain of SARS-CoV-2 nsp3, both across diverse coronaviruses, and across diverse SARS-CoV-2 samples. However, most of these variations are predicted to have little impact on ADPr binding. While this structural diversity is not favorable for the design of broad-spectrum ligands, competitive inhibitors mimicking interactions made by ADPr may have a broad spectrum of action.
References:
Schapira, M., Totrov, M., Abagyan, R. (1999). Prediction of the Binding Energy for Small Molecules, Peptides and Proteins. Journal of Molecular Recognition 12(3), 177-90 https://doi.org/10.1002/(SICI)1099-1352(199905/06)12:3<177::AID-JMR451>3.0.CO;2-Z
Grunewald, M. E., Chen, Y., Kuny, C., Maejima, T., Lease, R., Ferraris, D., Aikawa, M., Sullivan, C. S., Perlman, S., & Fehr, A. R. (2019). The coronavirus macrodomain is required to prevent PARP-mediated inhibition of virus replication and enhancement of IFN expression. PLOS Pathogens, 15(5), e1007756. https://doi.org/10.1371/journal.ppat.1007756
Michalska, K., Kim, Y., Jedrzejczak, R., Maltseva, N. I., Stols, L., Endres, M., & Joachimiak, A. (2020). Crystal structures of SARS-Cov-2 ADP-ribose phosphatase (ADRP): From the apo form to ligand complexes. https://doi.org/10.1101/2020.05.14.096081
