In my last post, I showed how we found the residues lining the ADP-ribose (ADPr) binding site of SARS-CoV-2 nsp3-Mac1 using its crystal structure (PDB: 6w02).
In this post, I will show the sequence diversity across UniProt entries from the Alpha- and Betacoronavirus genera and map that to the Mac1’s ADPr binding site using the same crystal structure (PDB: 6w02).
In the context of the emergence of future MERS-like or SARS-like coronaviruses from the bat strains circulating in bat reservoir species, it is imperative to do a broad survey of viral proteins to identify the best strategies for the development of broad-spectrum viral inhibitors. (Sheahan et al., 2020) For this exact reason, we are looking at twenty-seven reviewed sequences from Uniprot, where six belong to entries of Alphacoronavirus genus and twenty-one belong to the Betacoronavirus genus. We then assess their amino acid variability at the nsp3-Mac1 ADPr binding site (PDB: 6w02).
I made a diversity dendrogram focusing on the 29 sidechains surrounding the ADPr binding site of nsp3-Mac1. Shown below in figure 1 is the diversity dendrogram, where the consensus profile at each of the 29 sidechains is shown. Strikingly, only 11 out of 29 sidechains are conserved across these 27 entries: N37, A39, N40, H45, G48, A50, G97, S128, G130, I131, F132.

Figure 1. The diversity dendrogram of the ADPr-binding site of Mac1 for the entries of Alpha- and Betacoronavirus genera.
We made a diversity dendrogram of the 21 entries of the Betacoronavirus genus. As shown in figure 2, 14 out of 29 residues are conserved at Mac1 ADPr binding site: D22, N37, A39, N40, H45, G46, G48, A50, G97, L126, S128, G130, I131, F132.
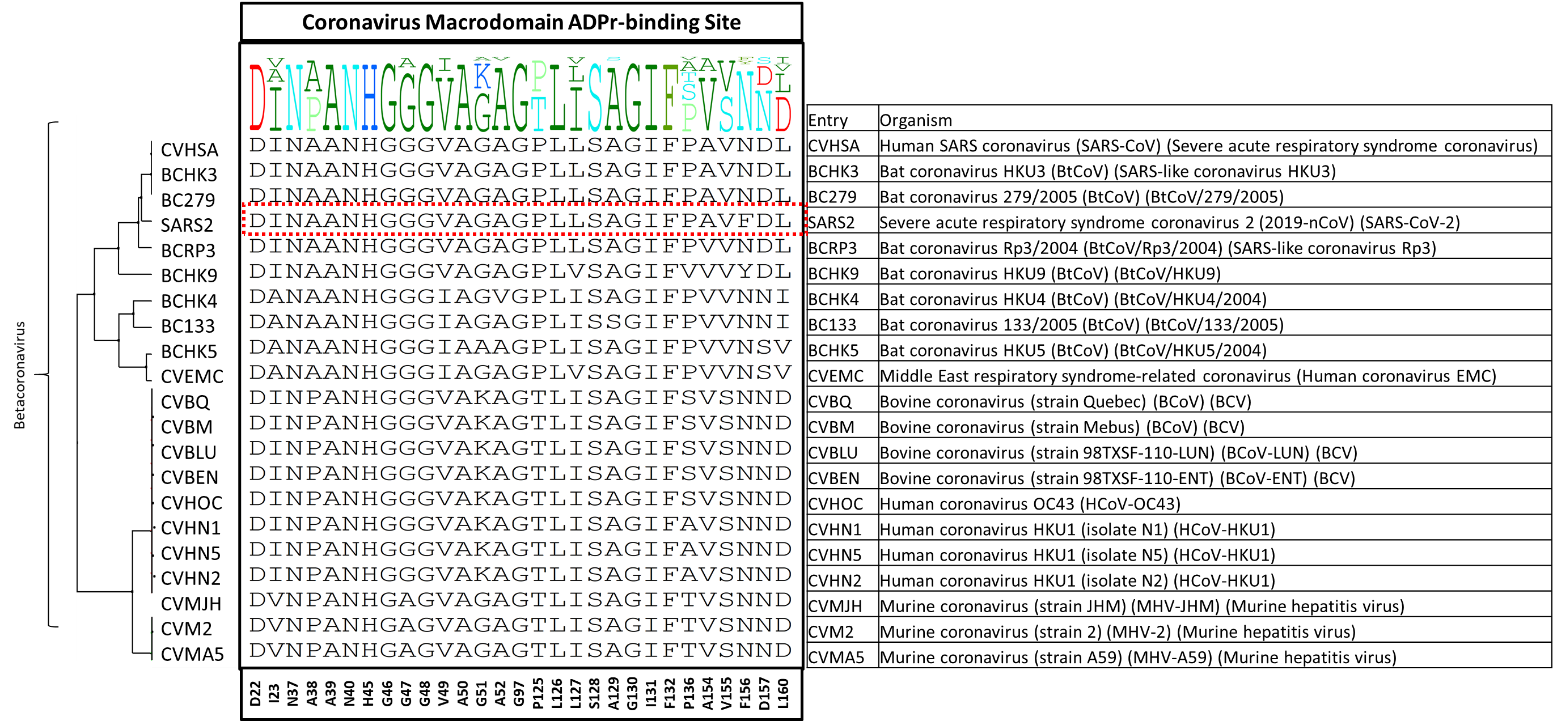
Figure 2. The diversity dendrogram of Mac1 ADPr binding site residues among the UniProt entries of the Betacoronavirus genus.
We mapped the variations observed among Alpha- and Betacoronavirus genera entries and the variations within only the Betacoronavirus entries onto the SARS-CoV-2 Mac1 crystal structure. The sidechains that are variable among the entries are highlighted in orange and the conserved sidechains are shown in green.
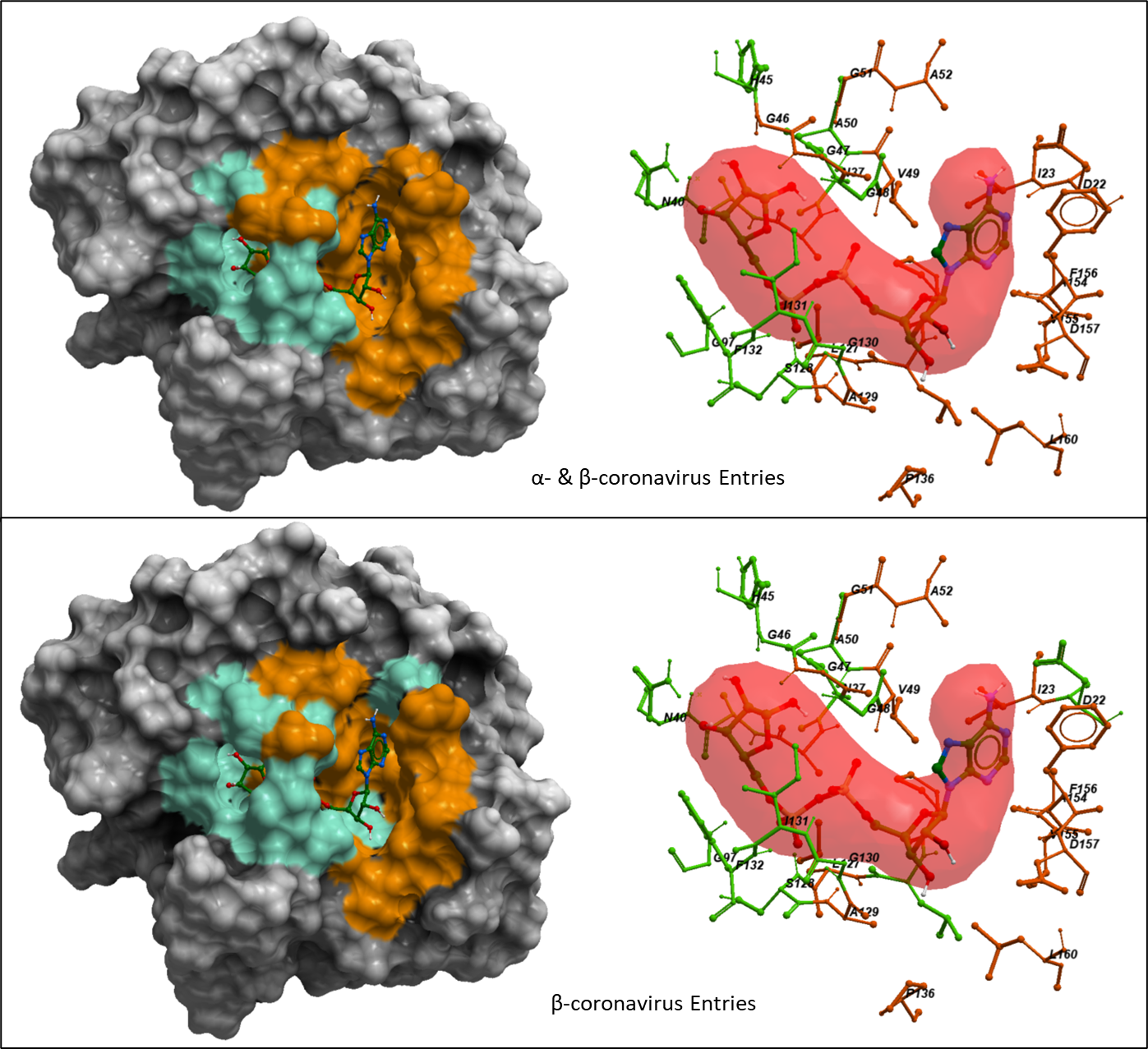
Figure 3. Mapping the genetic variation of UniProt entries belonging to Alpha- and Betacoronavirus genera (top) and among entries of Betacoronavirus genus (bottom) onto SARS-CoV-2 nsp3-Mac1 Crystal structure (PDB: 6w02). The green color shows the conserved residues and the orange color shows the non-conserved residues among the entries.
Based on figure 3, it is apparent that the residues lining the adenosine part of ADPr are not conserved (orange) while the residues surrounding the terminal ribose ring are conserved (green) among the entries of Alpha- and Betacoronavirus genera.
In one of my future posts, I will cover the predicted effect of each variation mentioned above in terms of how it alters the binding of the endogenous ligand, ADPr, to nsp3-Mac1.
To view the corresponding Zenodo report, please click here.
Stay in touch and leave your comments using the “Leave a comment” link at the top of this post. Stay Tuned for more updates on this project!
Reference:
Sheahan, T. P., Sims, A. C., Zhou, S., Graham, R. L., Hill, C. S., Leist, S. R., . . . Baric, R. S. (2020). An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 and multiple endemic, epidemic and bat coronavirus. Science Translational Medicine 12 (541). https://doi.org/10.1126/scitranslmed.abb5 883
