Background
GID4 is the substrate-recognition subunit of the CTLH E3 ubiquitin-protein ligase complex which has been shown to possess E3-ligase enzymatic activity (PIMID:31885576). GID4 was shown to bind proteins and peptides with a Pro/N-degron consisting of an unmodified N-terminal Pro and has the highest affinity for the peptide Pro-Gly-Leu-Trp (PIMID:29632410). In yeast Gid4 specifically targets the gluconeogenic enzymes (Fbp1, Icl1, and Mdh2) for proteasome-mediated degradation (PIMID: 31863529), however, substrate specificity for human GID4 has not been fully understood.
Assay validation
Cell-based thermal shift assays (CETSA) provide a native environment for measuring compounds’ target engagement and cell permeability. Standard western blot-based CETSA protocols are laborious, require high-quality antibodies to the target, and are generally not amenable to high-throughput compound screening. Therefore, we have adopted the HiBiT technology-based CETSA, which measures the thermal stability of a NanoLuc Luciferase fusion protein, to measure GID4 cellular target engagement in a 384-well plate format (Fig.1). The assay was validated with two compounds: 20964 and 12966, which showed significant dose-dependent stabilization of GID4 when tested in vitro by DSF. Using both compounds we observed reproducible stabilization of GID4 (Fig.2A) in cells. Since compound 20964 gave better Tm-shift (∆Tm), we tested if the stabilization is dose-dependent in a temperature range which gave the biggest difference between treated and untreated samples. Compound 20964 showed the dose-dependent stabilization of GID4 (Fig.2B). The effect was significant at 1 µM concentration (Fig.2C). These results validate the utility of the HiBiT based CETSA assay to measure cellular target engagement of GID4.
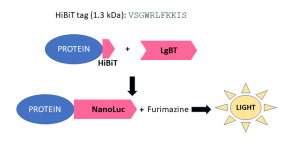
Fig.1. HiBIT protein tagging system. HiBiT is an 11 amino acid peptide tag that can be attached to the N or C terminus of the protein. The tag is detected with a reagent containing an inactive luciferase subunit, Large Bit (LgBiT), which rapidly binds to HiBiT to produce a functional NanoLuc luciferase enzyme that relies on the substrate furimazine to produce high intensity, glow-type luminescence.
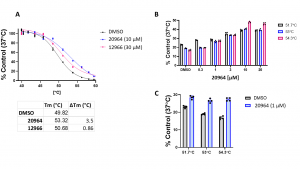
Fig.2. GID4 HiBiT CETSA target engagement assay validation. HeLa cells were transfected with N-terminally HiBiT-tagged GID4. Cells were treated with compounds and DMSO control for an hour before heating. After heating, cells were lysed and LgBiT and Luciferase substrate were added and the signal was measured. (A) Effect of 20964 (10 µM) and 12966 (30 µM) on the thermal stability of HiBiT-tagged GID4 relative to DMSO control. (B) 20964 showed the dose-dependent stabilization of GID4 in an indicated temperature range. (C) 20964 significantly stabilized GID4 at 1 µM in an indicated temperature range. The results are mean ± SEM of 4 technical replicates.
Experimental details can be found on Zenodo.
