by Mohammad Anwar Hossain1, Konstantin Popov2, Sumera Perveen3, Kesatebrhan Haile Asressu1, Kenneth Hugh Pearce Jr.2, Cheryl Arrowsmith3, Peter Brown1, Alexander Tropsha4, Tim Willson1
1Structural Genomics Consortium, The University of North Carolina at Chapel Hill, Chapel Hill, NC, USA; 2CICBDD, UNC Eshelman School of Pharmacy; 3The Structural Genomics Consortium, University of Toronto, Canada; 4CBMC, UNC Eshelman School of Pharmacy
Abstract:
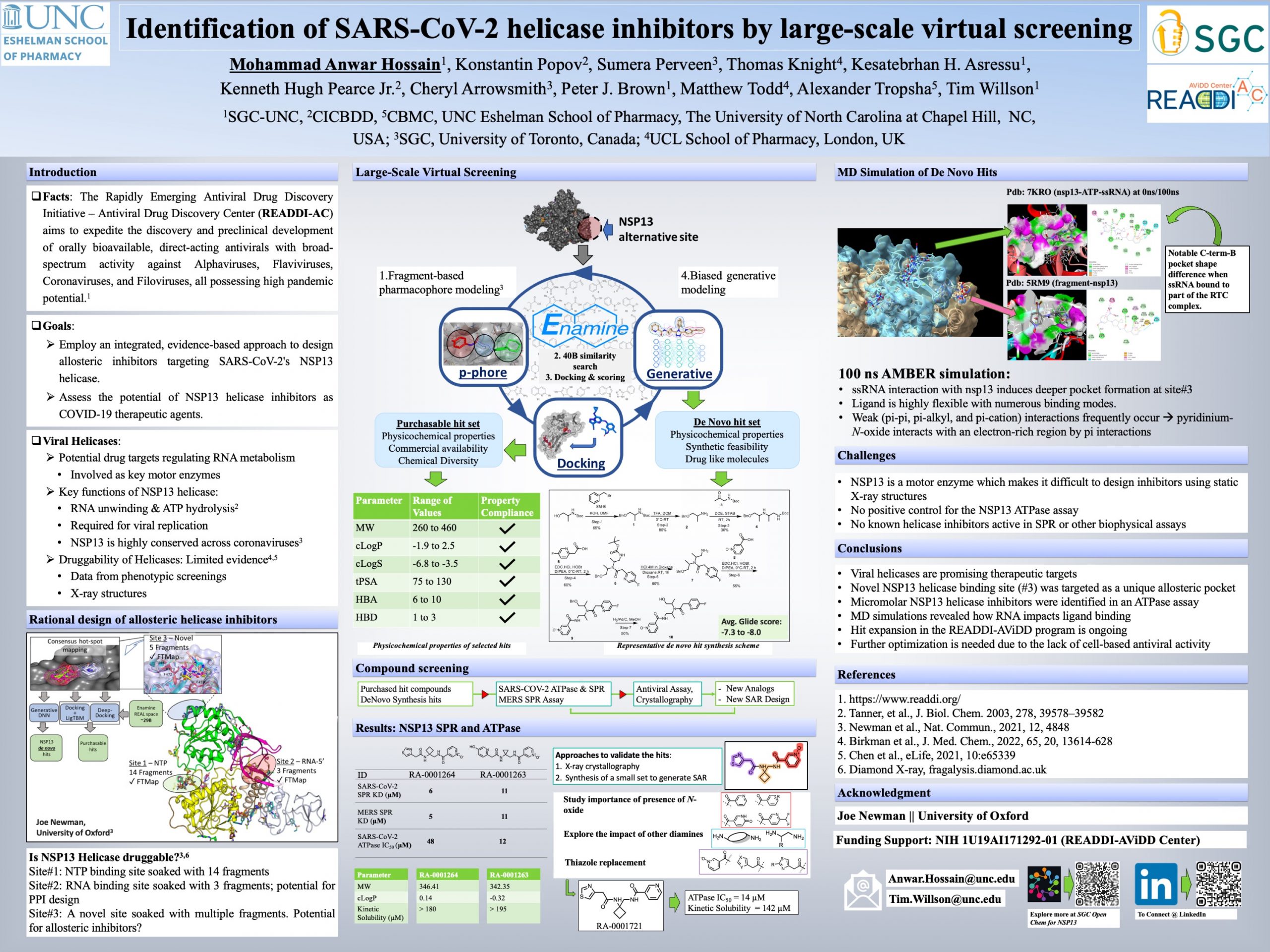
The COVID-19 pandemic, caused by the SARS-CoV-2 virus, exposed gaps in our nation’s preparedness for both the rapid development of antiviral drugs and the broader public health response. The high transmissibility of this novel pathogenic respiratory virus underscored the need for broad-spectrum antiviral drug development. The Rapidly Emerging Antiviral Drug Discovery Initiative AViDD Center (READDI-AC) was established to develop a pipeline of direct-acting antiviral hits, leads, and drug candidates with broad-spectrum activity against viruses of pandemic potential. SARS-CoV-2 NSP13 helicase is a target for the development of antivirals that may have activity on variants with resistance to protease inhibitors. Starting with a non-canonical binding pocket that was identified from the publicly available output of an NSP13 X-ray fragment screen, we employed a novel ultra-fast virtual screening methodology called Hit Discovery using Docking ENriched by GEnerative Modeling (HIDDEN GEM) to select de novo generated and purchasable small molecules. We selected 51 compounds from the Enamine REAL Space library and synthesized several additional exemplars of the de novo generated molecules. In total, 80 compounds were tested against NSP13 helicase using an ATPase assay and an SPR binding assay. Three N-oxide-containing compounds were identified with ATPase IC50 < 50 μM and SPR KD < 10 μM. Synthesis of additional analogs yielded a molecule with an IC50 = 14 μM in the ATPase assay. Additional characterization of this potential NSP13 inhibitor in orthogonal assays and by X-ray crystallography is in progress.
Poster PDF:GRC_Poster_final
Conference: Medicinal Chemistry Gordon Research Conference (GRC), New London, New Hampshire, Aug 2023
