PATL2: A Potential Contraceptive Target
A promising protein target for NHCs is Protein Associated with Topoisomerase II Homolog 2 (PATL2), a highly conserved, oocyte-specific mRNA-binding protein that represses translation1. For consanguineous families, the inheritance pattern for PATL2 is recessive; thus, infertility is often caused by homozygous mutations in the gene2. Although some mutations in PATL2 reduce expression levels, the molecular mechanism through which mutations lead to oocyte maturation arrest is not fully understood2–4. During oocyte growth, mRNA that is essential to maturation accumulates in the oocyte, in which about 30% of mRNA must be translationally repressed by proteins such as PATL2 until after fertilization5. PATL2 mRNA is expressed in immature oocytes with significant translation occurring during the primary follicle stage5. PATL2 concentration peaks during the secondary follicle stage, but the total amount of PATL2 increases as oocytes mature (Figure 1)5. In the absence of PATL2, genes involved in oocyte maturation and embryonic development are deregulated, demonstrating that PATL2’s function is critical to these processes5,6.
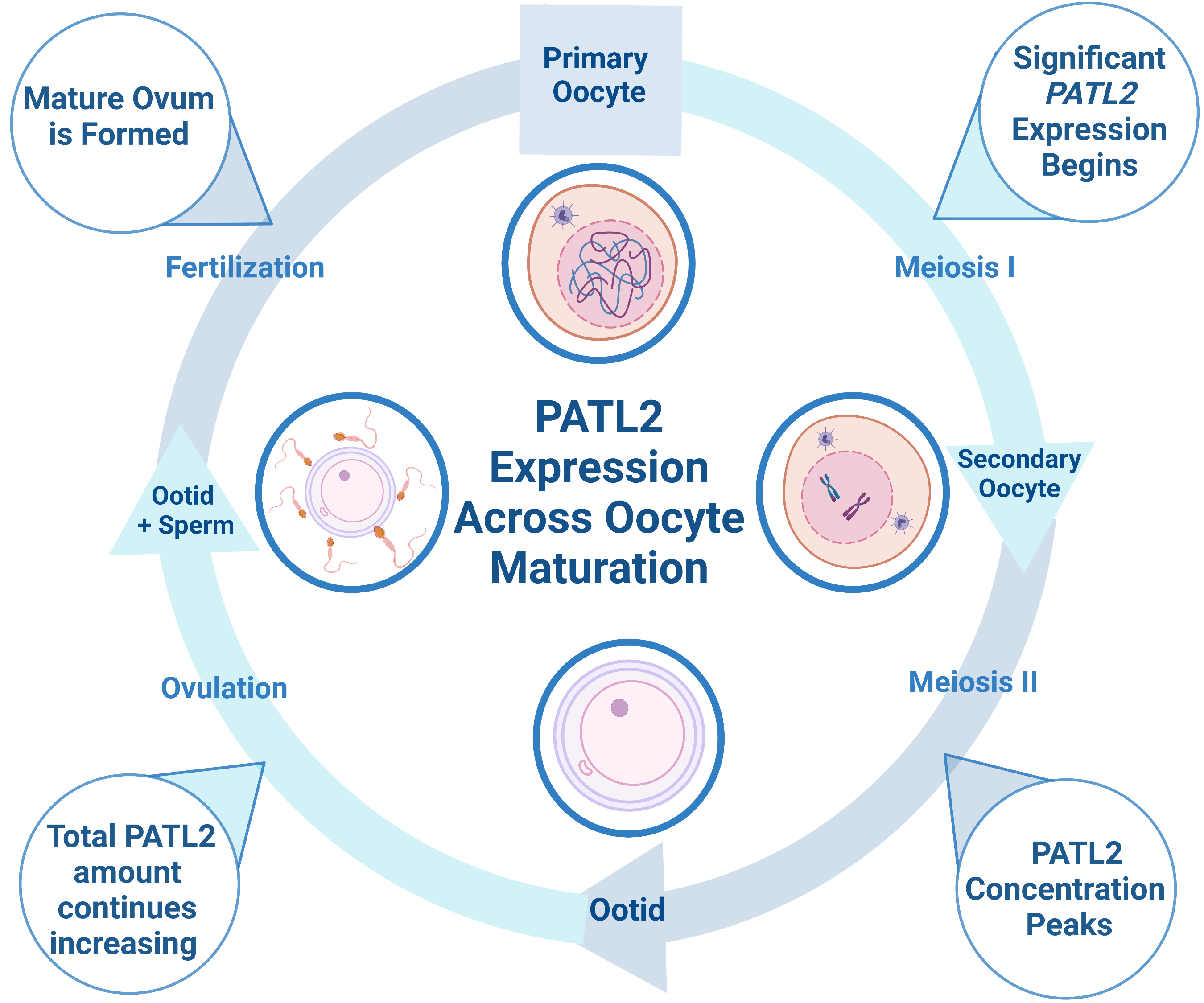
Figure 1: Diagram depicting PATL2 expression during oocyte maturation. PATL2 is expressed in immature oocytes, with significant translation starting at the primary oocyte stage. As oocytes mature, the total amount of PATL2 increases, but the PATL2 concentration peaks during the secondary follicle stage. Interestingly, PATL2 has a unique expression profile relative to other known RNA-binding proteins in oocytes. Both overexpression and the absence of PATL2 expression can cause oocyte maturation arrest. Figure made in bioRender.
PATL2 is a homolog of PATL1, which also contains a C-terminal PAT1 domain; the two proteins share 38% identity. PAT proteins contain a conserved N-terminal sequence, a Mid domain, a proline-rich region, and a C-terminal domain1,7. Within the C-terminus of PATL2, a PAT1 domain (amino acids 252-491) mediates PATL2’s mRNA-binding capabilities (Figure 2)2. In contrast to the C-terminus of PATL2, the N-terminal half (approximately 53% of the 543 amino acids) is predicted to be disordered (Figure 2). PATL1 has been shown to couple mRNA decapping and deadenylation in the 5’-3’ decay pathway through its interactions with the Lsm 1-7 complex8,9. PATL2 associates with some of the same proteins as PATL1, such as Lsm1 and Lsm4, of the Lsm1-7 complex, as well as Xrn19. Since PATL1 and PATL2 share sequence identity in the region of PATL1 that allows it to bind RNA, PATL2 likely retains similar RNA-binding properties1,10.
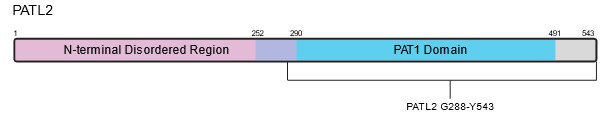
Figure 2: Schematic and predicted structure of human PATL2. A) Schematic depicting the relevant domains of PATL2. B) Predicted structure of full-length human PATL2 from AlphaFold2. The N and C-termini are labeled in black, along with amino acid G288, which serves as the start site for one of our PATL2 constructs. The disordered N-termini of the protein (aa 1-290) is colored magenta, while the ordered C-termini containing the PAT1 domain is colored blue (aa 291-543).
Proposed research:
The overarching goal of the proposed research is to enable drug discovery studies on PATL2. To this end, we will (1) structurally and biophysically characterize PATL2 interactions with RNA and protein binding partners (as this information is missing in the literature), (2) develop a biophysical assay (Protein-protein or protein-RNA interaction assays) suitable for compound screening and (3) identify small molecule binders that can be progressed to chemical probes to assess PATL2’s role in reproductive biology. To achieve these ends, PATL2, and various constructs of this protein, will be produced recombinantly and purified (Aim 1). Purified PATL2 will be used to characterize its RNA-binding abilities, protein-protein interactions, translation repression, and other biophysical properties (Aim 2). Several of the planned constructs are also suitable for X-ray crystallography, allowing us to structurally elucidate the PATL2 interaction with its binding partners (Aim 3). Finally, we will combine the knowledge gained from Aims 1-2 to screen for small molecules capable of inhibiting the PATL2’s RNA-binding or protein-binding functions (Aim 4).
Aim 1: Recombinant Expression and Purification of PATL2 Constructs
Several PATL2 constructs for recombinant expression and purification were designed using the AlphaFold2 program to analyze the predicted structure of PATL2 and design potential domain boundaries for protein expression constructs. Expression constructs were first tested on a small scale to determine which constructs could generate milligram quantities of soluble protein. From the constructs tested, we were able to successfully purify several PATL2 constructs. We first purified an N-terminally tagged hexahistidine protein construct consisting of amino acids G288-Y543 from the SF9 insect cell/baculovirus system, however the yield was low (0.04 mg/liter of cells). When purifying the same construct from E. coli, our yield was tenfold higher. Both constructs were purified using nickel affinity chromatography, followed by reverse nickel affinity, and finally, gel filtration (Figure 3).
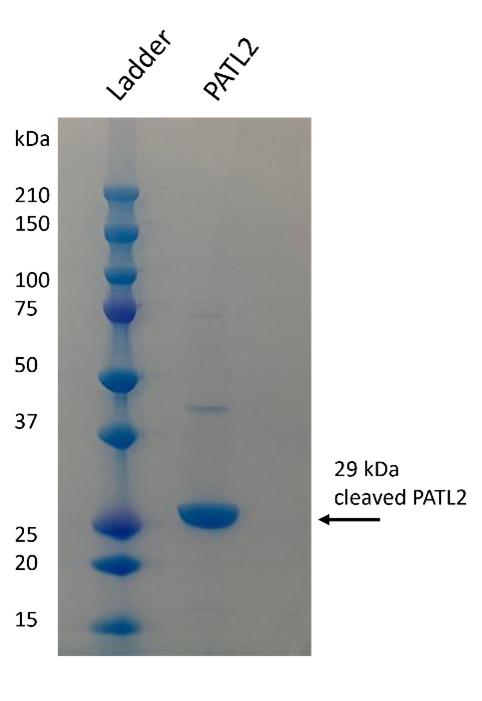
Figure 3: Purified PATL2 G288-Y543. The SDS-PAGE gel shows the purified protein. Protein identity was confirmed by mass spectrometry. Protein elutes as a monomer on gel filtration column.
Aim 2: Biophysical Assays to Explore PATL2’s Function
PATL2 regulates specific genes implicated in oocyte maturation and early embryonic development as opposed to globally repressing translation, suggesting that PATL2 has sequence or structure-specific binding properties5. This hypothesis is supported by the observation that both human PATL1 and Xenopus PATL2 were found to bind GQs, as well as poly(G) and poly(U) homopolymers, in vitro10. Based on these findings, we purchased the synthetic RNA GQ from the 5’ UTR of the NRAS proto-oncogene used in this publication. We also created a mutant version that would be incapable of forming GQs. Using fluorescence anisotropy, we observed only very weak binding by our PATL2G288-Y543 construct to either RNA. Our findings suggest that these RNAs may not be the target of Human PATL2, or perhaps the N-terminus of PATL2 is needed for binding to GQs. Moving forward, we will (1) attempt to purify a construct of PATL2 that contains the N-terminus, (2) test if PATL2 interacts with RNA as a part of a complex with other proteins (such as those mentioned below), and (3) we may perform RNA pull-down assays to identify RNA targets specifically of Human PATL2.
Likewise, it is a priority to examine PATL2 interaction partners and verify if PATL2 directly binds these proteins in vitro. Using coimmunoprecipitation (co-IP) experiments, previous studies have identified Lsm1, Lsm4, and Xrn1 as PATL1 and PATL2 interactors.9,11. LSM1 and LSM4 have been successfully purified for Biolayer Interferometry assays to determine if PATL2 directly binds either of these proteins in vitro. The aforementioned co-IP experiments were performed with HEK293 cells as opposed to oocytes, where PATL2 is highly expressed. Therefore, we plan to ship our purified PATL2 for antibody generation, as antibodies will be needed for our co-IP assays in oocytes. In the meantime, we are currently in the process of testing if the LSM1-7 complex will co-precipitate with full-length PATL2, or with the PAT1 domain from SF9 cells. Additionally, Mario Bengtson’s lab is cloning PATL2 into a mammalian expression vector to identify additional PATL2 interaction partners.
Aim 3: Structural Characterization of PATL2 through X-ray Crystallography
To date, the experimental structure for human PATL2 has not been reported. As the PAT1 domain of PATL1 has been crystallized, there is a high probability that we will can crystallize this domain from PATL2 in order to support structure-guided drug discovery7. We set up initial crystal screens with the PAT1 domain of PATL2 yet were unsuccessful in generating crystals. We are currently testing if proteolytic digestion of the PAT1 domain will provide us with a smaller domain that is more amenable to crystallization. As proteins are often stabilized by their interaction partners, we will attempt to co-crystallize a complex of PATL2 with its binding partners, which would contribute greatly to the field’s knowledge of this protein. Moreover, this structure would enable us to design evidence-based molecules capable of disrupting PATL2’s interactions that promote reproduction.
Aim 4: Test the Ability of Small Molecules to Inhibit PATL2 Function
Once we have developed a fuller understanding of the functions of PATL2 from Aims 2 and 3, we will design assays to screen for compounds that inhibit the protein’s RNA-binding or protein-binding function. As alluded to earlier, we will benefit from knowing which regions of PATL2 contribute to its various functions. This will allow us to pinpoint regions of the protein to target for protein degradation or small molecule inhibitors. Another approach to determine how to target PATL2 will be analyzing the PATL2 mutants that result in infertility. Several PATL2 mutations have been identified, and work in previous aims will explore how these mutations alter PATL2’s ability to bind to RNA and to its protein interaction partners. This information will inform our decision on small molecules to test, and suitable assays to perform.
Acknowledgements
I would like to acknowledge Alma Seitova, Ashley Hutchinson, Maria Kutera, and Peter Loppnau for their contributions to cloning and expressing PATL2 protein constructs. I would like to thank Hong Zeng for her work to purify some of the PATL2 constructs, and Mario Bengtson for his lab’s collaboration. I would also like to acknowledge Levon Halabelian and Aled Edwards for their input and supervision of this work.
This project is funded by the Bill and Melinda Gates Foundation. The Structural Genomics Consortium is a registered charity (no: 1097737) that receives funds from AbbVie, Bayer AG, Boehringer Ingelheim, Genentech, Genome Canada through Ontario Genomics Institute [OGI-196], the EU and EFPIA through the Innovative Medicines Initiative 2 Joint Undertaking [EUbOPEN grant 875510], Janssen, Merck KGaA (aka EMD in Canada and US), Pfizer, Takeda and the Wellcome Trust [106169/ZZ14/Z].
References
(1) Maddirevula, S.; Coskun, S.; Alhassan, S.; Elnour, A.; Alsaif, H. S.; Ibrahim, N.; Abdulwahab, F.; Arold, S. T.; Alkuraya, F. S. Female Infertility Caused by Mutations in the Oocyte-Specific Translational Repressor PATL2. Am. J. Hum. Genet. 2017, 101 (4), 603–608. https://doi.org/10.1016/j.ajhg.2017.08.009.
(2) Liu, Z.; Zhu, L.; Wang, J.; Luo, G.; Xi, Q.; Zhou, X.; Li, Z.; Yang, X.; Duan, J.; Jin, L.; Zhang, X. Novel Homozygous Mutations in PATL2 Lead to Female Infertility with Oocyte Maturation Arrest. J. Assist. Reprod. Genet. 2020, 37 (4), 841–847. https://doi.org/10.1007/s10815-020-01698-6.
(3) Huang, L.; Tong, X.; Wang, F.; Luo, L.; Jin, R.; Fu, Y.; Zhou, G.; Li, D.; Song, G.; Liu, Y.; Zhu, F. Novel Mutations in PATL2 Cause Female Infertility with Oocyte Germinal Vesicle Arrest. Hum. Reprod. 2018, 33 (6), 1183–1190. https://doi.org/10.1093/humrep/dey100.
(4) Wu, L.; Chen, H.; Li, D.; Song, D.; Chen, B.; Yan, Z.; Lyu, Q.; Wang, L.; Kuang, Y.; Li, B.; Sang, Q. Novel Mutations in PATL2: Expanding the Mutational Spectrum and Corresponding Phenotypic Variability Associated with Female Infertility. J. Hum. Genet. 2019, 64 (5), 379–385. https://doi.org/10.1038/s10038-019-0568-6.
(5) PATL2 Is a Key Actor of Oocyte Maturation Whose Invalidation Causes Infertility in Women and Mice. EMBO Mol. Med. 2018, 10 (5), e8515. https://doi.org/10.15252/emmm.201708515.
(6) Chen, B.; Zhang, Z.; Sun, X.; Kuang, Y.; Mao, X.; Wang, X.; Yan, Z.; Li, B.; Xu, Y.; Yu, M.; Fu, J.; Mu, J.; Zhou, Z.; Li, Q.; Jin, L.; He, L.; Sang, Q.; Wang, L. Biallelic Mutations in PATL2 Cause Female Infertility Characterized by Oocyte Maturation Arrest. Am. J. Hum. Genet. 2017, 101 (4), 609–615. https://doi.org/10.1016/j.ajhg.2017.08.018.
(7) Braun, J. E.; Tritschler, F.; Haas, G.; Igreja, C.; Truffault, V.; Weichenrieder, O.; Izaurralde, E. The C-Terminal Alpha-Alpha Superhelix of Pat Is Required for MRNA Decapping in Metazoa. EMBO J. 2010, 29 (14), 2368–2380. https://doi.org/10.1038/emboj.2010.124.
(8) Sharif, H.; Conti, E. Architecture of the Lsm1-7-Pat1 Complex: A Conserved Assembly in Eukaryotic MRNA Turnover. Cell Rep. 2013, 5 (2), 283–291. https://doi.org/10.1016/j.celrep.2013.10.004.
(9) Ozgur, S.; Chekulaeva, M.; Stoecklin, G. Human Pat1b Connects Deadenylation with MRNA Decapping and Controls the Assembly of Processing Bodies. Mol. Cell. Biol. 2010, 30 (17), 4308–4323. https://doi.org/10.1128/MCB.00429-10.
(10) Marnef, A.; Maldonado, M.; Bugaut, A.; Balasubramanian, S.; Kress, M.; Weil, D.; Standart, N. Distinct Functions of Maternal and Somatic Pat1 Protein Paralogs. RNA 2010, 16 (11), 2094–2107. https://doi.org/10.1261/rna.2295410.
(11) Szklarczyk, D.; Gable, A. L.; Nastou, K. C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N. T.; Legeay, M.; Fang, T.; Bork, P.; Jensen, L. J.; von Mering, C. The STRING Database in 2021: Customizable Protein-Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49 (D1), D605–D612. https://doi.org/10.1093/nar/gkaa1074.
