My previous results showed that overexpression of HDAC11 decrease basal acetylation levels of exogenous histones as well as decrease the acetylation increase caused by histone acetyltransferases. In order to determine if the observed effect is caused by catalytic activity of HDAC11, I used catalytic HDAC11 mutants Y304H and H142,143A (PIMID: 30819897, 28927261) and published HDAC11 inhibitors SIS17, SIS7 (25 µM, PMID:31264832) and FT895 (5 µM, PMID: 29776742) for validation. I run the assay in different cell lines (HEK293T, Hela and MCF7) with various amounts of HDAC11 and HATs (GCN5, MYST3), various assay duration (1-2 days) and looked at the changes in H3KAc, H3K18Ac, H3K27Ac, H3K9Ac levels by western blot.
Summary
- The results are highly variable mostly due to low signal and effect of HDAC11 overexpression on cell growth (less cells).
- In most conditions tested HDAC11 catalytic mutants showed similar decrease in acetylation compared to WT HDAC11.
- None of the compounds inhibited the decreased acetylation caused by HDAC11 overexpression. Compound FT895 showed toxic effect even at 5 μM.
- The results indicate that the decrease in histone acetylation levels caused by HDAC11 overexpression does not depend on HDAC11 catalytic activity
Here is the representative graph form one of the experiments.
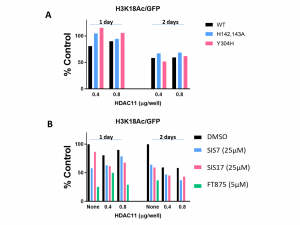
HDAC11 cellular assay validation with catalytic mutants and inhibitors.
Hela cells were co-transfected with GFP-tagged histone H3 and or with different amounts of Flag-tagged HDAC11 (Wt or Y304H and H142,143A catalytic mutant) or empty vector and treated with inhibitors for 1 or 2 days. https://zenodo.org/record/3525243#.XbxdiDNKg2w
