Molecular dynamic simulations are a powerful tool for modeling biomolecular systems such as ligand binding to proteins. In a previous post I was able to get co-crystal structures of ligands which bind to the USP5 zinc finger ubiquitin binding domain (Zf-UBD). In order to explore the structure activity relationship (SAR) of the chemical series, so I can rationally design more potent ligands, I used MolPort and Emoleules to find chemically related, commercially available molecules, and then used free energy perturbation (FEP) to prioritize compound ordering. You can see full experimental details on Zenodo. Free energy perturbation (FEP) is a computational chemistry tool that allows for the thermodynamic calculation of relative binding free energy1. The binding free energy difference can be calculated by the difference of the free energy of transforming ligand 1 to ligand 2 in the solvent and in complex with a receptor (Figure 1)1.
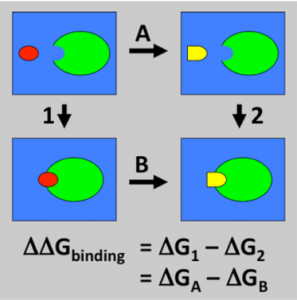
Figure 1. Thermodynamic pathway of calculating the free energy difference1
As an example, for DAT180, I did a substructure search using constraints on the aromatic rings as well as the carboxyl group of the compound which was shown to be an important interaction in the co-crystal structure. I got a few hundred hits which I docked to the Zf-UBD while imposing H-bonding constraints between the R221/Y261, and the carboxylate of the compound, important interactions in the binding pocket (Figure 2).
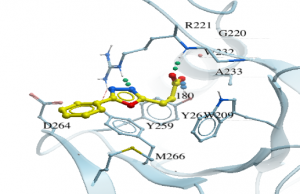
Figure 2. Co-crystal structure of USP5 Zf-UBD and DAT180 (PDB: 6DXT)
After docking, 30 compounds were selected on the basis of visual inspection & chemistry intuition, molecular weight, and docking scores. I used FEP as a tool to estimate the free energy difference of analogues of the ligands from the co-crystal structures that bind with micromolar affinity, as shown by SPR, for a final selection of 17 compounds (Figure 3).
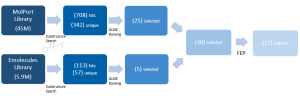
Figure 3. Hit expansion summary of DAT180
I’ve ordered the selected compounds from the substructure searches/FEP. Hopefully, the next batch of compounds I test experimentally will have increased potency. More on this soon!
1Acc. Chem. Res., 2017, 50 (7), pp 1625–1632
