In vivo characterization of antibodies directed against TREAT-AD target proteins in mouse model of AD pathology
Suzanne Doolen, Riham Ayoubi, Carl Laflamme, Ranjita Betarbet, Elizabeth Zoeller, Sean-Paul G. Williams and Stacey J. Sukoff Rizzo
https://doi.org/10.5281/zenodo.6612215
Our goal in support of the Emory-Sage-Structural Genomics Consortium (SGC) TREAT-AD Center is to develop and identify high-quality tools to test target or mechanistic hypotheses in animal models. As commercial research antibodies become available and validated in commercially available knock-out cell lines, it is essential to extend the characterization of these antibodies in animal models. This allows not only for confirmation of the presence and/or changes in the protein level of target proteins but will also potentially serve as a reagent for future in vivo target engagement studies. Here we have used the 5xFAD mouse to characterize target proteins. While the 5xFAD is a well-characterized and highly published transgenic model that manifests Aβ plaque deposition as early as 4-6 months of age [1, 2], we have not previously characterized the expression of new target proteins being pursued by the TREAT-AD center. Here we have characterized the expression of Moesin, CD44, Midkine and SFRP1 in the 5xFAD mouse model.
YCharOS (https://ycharos.com/) is an open-science organization which has industrialized an antibody characterization platform [3] formalized at the Montreal Neurological Institute [4]. Together with YCharOS, we have characterized commercial antibodies against Moesin [5], CD44 [6], Midkine [7] and SFRP1 [8] using corresponding human knockout cell lines. We next have used the best performing antibodies on protein extracts from cortex of 9 month aged 5xFAD mouse brain homogenates and compared to age-matched C57BL/6J controls. Our immunoblotting results depicting their reactivity in mouse brain homogenate are shown in Figure 1 (higher resolution image available at Zenodo). The antibody source information and the fold change in protein levels in 5xFAD are in Table 1. In Figure 1A anti-Moesin ab52490 reacted in mouse brain homogenate with a predicted molecular weight of 68 kD. Moesin protein expression was 1.96 times higher in 5xFAD compared to WT mouse brain. In Figure 1B anti-CD44 ab189524 reacted with a band at the predicted size of 82 kD. CD44 protein expression was 2.89 times higher in 5xFAD compared to WT mouse brain. Anti-Midkine AF7769 reacted with a band ~16 kD and a 24.7 times greater expression in 5xFAD compared to WT mouse brain (Figure 1C). Anti-SFRP1 ab267466 reacted with a band at 35 kD as predicted. SFRP1 protein expression was 5.96 times greater in 5xFAD compared to WT mouse brain (Figure 1D).
Studies of the human AD brain proteome revealed modules of proteins that were strongly correlated with higher neuropathological burden and worse cognitive outcomes [9, 10]. One such module (M4, astrocyte/microglia metabolism module), includes Moesin and CD44 and has the strongest correlation with higher neuropathological burden and worse cognitive outcomes [9, 10]. Module 4 (M4) is enriched in microglial and astrocytic proteins and contains proteins involved in response to inflammation. Another novel AD-associated Module, M42 or the matrisome module, which has a correlation of 0.75 with global pathology, and as such a source of promising therapeutic targets and biomarkers for AD is of significant interest. Midkine and SFRP1 belong to M42 [11]. It is of interest and significance that the increased expression of these proteins, i.e. Moesin, CD44, Midkine and SFRP1 in the 5xFAD mice model of AD, that are elevated in AD patient brain samples [10, 11], has also been confirmed by TMT proteomics by the Seyfried lab recently (unpublished data).
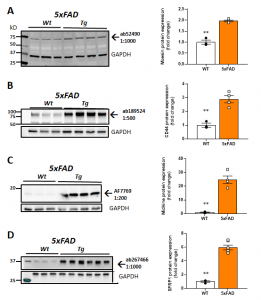
| Fig 1. Validation of target protein expression in AD mouse brain as a model system for potential application for in vivo target engagement studies. Characterization in 8-9 month 5XFAD mice. Representative western blots (left) and quantification (right) using the following antibodies: A. Anti-Moesin; (Abcam #ab52490) B. Anti-CD44 (Abcam #ab189524). C. Anti-midkine (R&D Systems #AF7769). D. Anti-SFRP1 (Abcam #ab267466). **P < 0.01 by t-test. See higher resolution image at Zenodo. |
Table. 1. Summary of commercial antibodies use in western blot assays.
| Target protein | Product # | Company | Source | Dilution used | RRID (Antibody Registry) | Clonality | Clone # | Conc. (μg/μL) | Fold change in 5xFAD brain |
| Moesin | ab52490 | Abcam | Rabbit | 1:1000 | AB_881245 | recombinant-mono | EP1863Y | 0.19 | 1.96 |
| CD44 | ab189524 | Abcam | Rabbit | 1:500 | AB_2885107 | recombinant-mono | EPR18668 | 0.47 | 2.89 |
| Midkine | AF7769 | R&D Systems | Sheep | 1:200 | AB_291796 | polyclonal | – | 1.0 | 24.7 |
| SFRP1 | ab267466 | Abcam | Rabbit | 1:1000 | AB_2904616 | recombinant-mono | EPR23092-253 | 0.46 | 5.96 |
Methods
All animal studies were reviewed and approved by the University of Pittsburgh Institutional Animal Care and Use Committee prior to study initiation. Adult male and female 5xFAD (JAX MMRRC Stock #034840) and C57BL/6J (JAX stock #000664) were received from The Jackson Laboratory and group-housed within sex (up to 4 per cage). For terminal tissue collection, mice were anesthetized with isoflurane anesthesia and brains were collected following decapitation. The brain was extracted and rinsed in ice-cold PBS, cerebellum was removed, and the cortex was bisected into left and right hemispheres then snap frozen and stored at -80 C until use. Each hemibrain was weighed prior to homogenizing on ice in tissue homogenization buffer (THB; 2 mM Tris (pH 7.4), 250 mM sucrose 0.5 mM EDTA 0.5 mM EGTA) supplemented with Pierce Protease Inhibitor (Thermo Scientific #A32953) and Phosphatase Inhibitor Cocktail 2 (Sigma-Aldrich #P5726 St. Louis, MO). Total protein concentration was measured using a Bradford assay [12]. Equal amounts of protein (25 μg) were separated with SDS-polyacrylamide gel electrophoresis (4–15% Mini-PROTEAN® TGX™ Precast Protein Gels, 10-well, 30 µl, BIORAD Cat#4561083) and transferred onto a nitrocellulose membrane. Non-specific binding was blocked using EveryBlot Blocking Buffer (BIORAD Cat#12010020) for 5 min at room temperature. Primary antibodies were prepared in EveryBlot Blocking buffer at the dilutions indicated in Table 1. Blots were immersed in primary antibody solutions overnight at 4 C with gentle rocking. Membranes were then washed with 1X TBST (3 × 5 min) and immersed in 1:1000 fluorescent secondary antibody (StarBright blue 700 goat anti-rabbit IgG, BIORAD Cat#12004161) and 1:2000 hFAB™ Rhodamine Anti-GAPDH Antibody (BIORAD Cat#12004167) for 1 h at room temperature. Membranes were then washed (2 × 5 min in 1X TBST; 2 × 5 min in 1X TBS) and scanned with a BIORAD ChemiDoc MP Imaging System. Pixel intensity was quantified using ImageJ the public domain NIH Image program (available at http://rsb.info.nih.gov/nih-image/). Statistical analyses and graphing were performed using GraphPad Prism Version 9.3.1 (San Diego, CA).
Acknowledgements
This work was supported by the NIH National Institutes on Aging TREAT-AD Center for Alzheimer’s Disease grant U54AG065187.
- Jawhar, S., et al., Motor deficits, neuron loss, and reduced anxiety coinciding with axonal degeneration and intraneuronal Abeta aggregation in the 5XFAD mouse model of Alzheimer’s disease. Neurobiol Aging, 2012. 33(1): p. 196 e29-40 DOI: 10.1016/j.neurobiolaging.2010.05.027.
- Oakley, H., et al., Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J Neurosci, 2006. 26(40): p. 10129-40 DOI: 10.1523/JNEUROSCI.1202-06.2006.
- Laflamme, C., et al., Implementation of an antibody characterization procedure and application to the major ALS/FTD disease gene C9ORF72. Elife, 2019. 8 DOI: 10.7554/eLife.48363.
- Laflamme, C., et al., Opinion: Independent third-party entities as a model for validation of commercial antibodies. N Biotechnol, 2021. 65: p. 1-8 DOI: 10.1016/j.nbt.2021.07.001.
- Alshafie, W.a., et al., Antibody Characterization Report for Moesin. 2021 DOI: 10.5281/zenodo.4724169.
- Ayoubi, R.a., et al., Antibody Characterization Report for CD44 antigen. 2021 DOI: 10.5281/zenodo.4730966.
- Ayoubi, R.a., P.S.a. McPherson, and C. Laflamme, Antibody Characterization Report for Midkine. 2021 DOI: 10.5281/zenodo.5644321.
- Ayoubi, R.a., P.S.a. McPherson, and C. Laflamme, Antibody Characterization Report for Secreted frizzled-related protein 1. 2022 DOI: 10.5281/zenodo.6370454.
- Seyfried, N.T., et al., A Multi-network Approach Identifies Protein-Specific Co-expression in Asymptomatic and Symptomatic Alzheimer’s Disease. Cell Syst, 2017. 4(1): p. 60-72 e4 DOI: 10.1016/j.cels.2016.11.006.
- Johnson, E.C.B., et al., Large-scale proteomic analysis of Alzheimer’s disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat Med, 2020. 26(5): p. 769-780 DOI: 10.1038/s41591-020-0815-6.
- Johnson, E.C.B., et al., Large-scale deep multi-layer analysis of Alzheimer’s disease brain reveals strong proteomic disease-related changes not observed at the RNA level. Nat Neurosci, 2022. 25(2): p. 213-225 DOI: 10.1038/s41593-021-00999-y.
- Bradford, M.M., A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem, 1976. 72: p. 248-54 DOI: 10.1006/abio.1976.9999.
