The goal of the Structural Genomics Consortium (SGC) is to discover and share selective small molecule inhibitors of protein kinases. Kinases have key roles in cell signaling, regulation of cell cycle progression, metabolism and other significant biological function. Cancers, immunological and metabolic diseases, among other ailments are caused by deregulation of kinase function. Protein kinases are now considered an extremely important group of drug targets, and their inhibitors can have life-saving therapeutic value. In 2001, Imatinib was the first kinase inhibitor was approved for market. Since then over 38 have been approved, with many more in various phases of clinical testing.
My aim within SGC is to develop a selective CaMKK2 chemical probe which will have suitable properties for in vivo testing. CaMKK2 is a serine/threonine kinase. It downstream signaling activates CaMK1, CaMK4 and AMPK, which in turn regulate several distinct and important biological responses. CaMKK2 deregulation is implicated in prostate cancer, breast cancer and hepatic cancer, showing CaMKK2 to be an important target of which selective inhibitors are promptly needed.
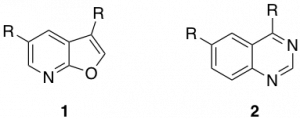
Excellent work towards these inhibitors has already been carried out by the SGC. These inhibitors have been mainly based around the furopyridine (1) and quinoline (2) cores.
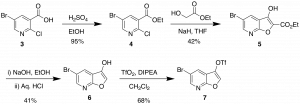
The route to the furopyridine core is described in Scheme 1. It generates the furopyridine 7 that has 2 functional handles which can be utilized in a variety of couplings. Under the right conditions there is selective functionalization of the triflate which we have taken advantage of in the library synthesis.
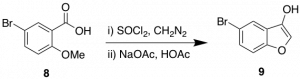
In my time with the SGC so far, I have utilized this route to begin generating a library of furopyridine analogues. Although the synthesis to key compound 7 is only 4 steps the yields do mean a significant loss in material. In an attempt to expedite access to 7, I will attempt a synthesis using Arntd-Eistert chemistry. This chemistry has been effectively applied in the synthesis of benzofurans previously as shown in Scheme 2.
In my next post I will describe the results of this synthesis, the analogues synthesized so far and any new biological data that has been collected.
