I decided to test a set of my ALK2 GS loop phosphomutants for activity. As I’ve written about previously, I needed to optimise the concentrations of receptors and decided that 60uM was the right concentration to use as showing activity with the mutants and also being workable in the assay.
I set up three different ALK2 mutants alongside the wild type ALK2 to compare them and used ACVR2 as my necessary type II receptor for SMAD1 activation.
I took time points at 0, 1, 5, 15, 30, 45 min, 1h, 1h15, 1h30, 1h45, 2h. Taking 1ul of sample and mixing it with 60ul of mass spec buffer (water + 0.1% formic acid)
I ran these on the mass spec to check for and monitor activity on all components but regrettably ended up seeing zero activity on SMAD1 even after 2h in the wildtype positive control. I did see phosphorylation on the type one receptors but nothing on the SMAD1 which suggested to me that something had gone horribly wrong with the type II receptor and led to it being unable to facilitate SMAD1 activation (as has been previously established to be necessary).
This was, to put it mildly, very frustrating.
So I went and purified a fresh stock of ACVR2 and did a small scale test using just the wild type to see if the positive control worked. This time it did so I set up a larger scale experiment using all the constructs we’ve managed to purify so far (which is all of those created so far except c107, c111 and c112).
This time we saw phosphorylation on SMAD and after 2h we could see strong signal and still make out the differences between the activity of the constructs and so further analysis was done on the 2h time point.
Differences between the different mutants stood out instantly – none of them increased the activity compared to the wild type but there were distinct effects on both the SMAD1 and ALK2 auto-phosphorylation patterns.
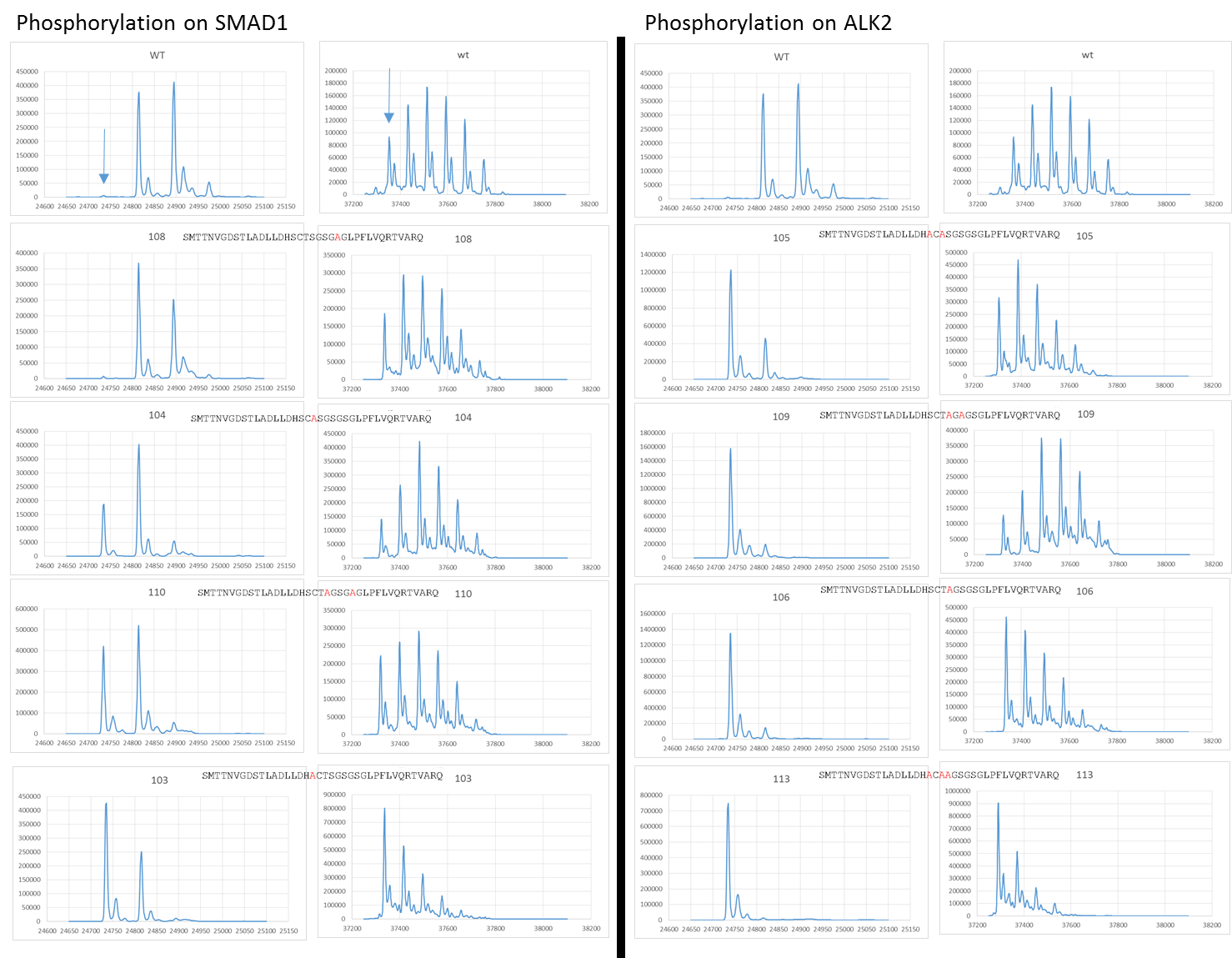
Summary figures showing the phosphorylation of SMAD1 and ALK2 using various ALK2 GS loop mutants. The top row on both columns shows the wild type positive control for comparison. The constructs are then ranked from least to most effected moving down the first column (with matching SMAD1 and ALK2 traces) and then down the second column.
The last four were the most extreme in terms of knocking out SMAD phosphorylation with the fourth leading to very low levels of phosphorylation and last three basically completely stopping it. When we look at the mutations these four constructs contain we can see a theme in common.
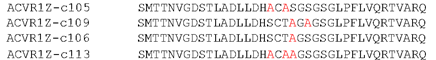
Figure showing the mutations contained within the four most detrimental mutations.
The three most detrimental constructs (c109, c106 and c113) all contained a S190A mutation with the fourth (c105) containing a T189A mutation just prior to the S190 residue. This suggests that S190 has a key role in facilitating SMAD1 phosphorylation.
Next thing to test is the constructs I’m missing (they need expressing and purifying) and to repeat this experiment before also running the results on a western blot for confirmation.
If you’d like more details then please do visit over on Zenodo.
