Hi,
Recently, I have started working on NanoBret PPI technology for chromatin factors and E3 ligases are here is one of the assays I developed.
Background (source: PMID28179136).
The repressive Nucleosome Remodeling and histone Deacetylation (NuRD) complex alters chromatin structure by coupling ATP-dependent remodeling activity with histone deacetylation function. It is composed of six different types of subunits: Metastasis Associated (MTA) proteins MTA1/2/3, HDAC1/2, histone chaperone RBBP4/7 forming a complex, as well as CpG-binding protein MBD2/3, p66α/β and includes the peripheral association of the ATP-dependent helicase CDH3/4. RBBP4 and RBBP7 are seven-blade β-propeller WD40 proteins that bind MTA through the same surface as histone H4, and therefore RBBP4/7 only bind histone H3 and not H4 in the complete NuRD complex. The Stoichiometry of the MTA1-RBBP4 complex is 2:4 (Fig 1).
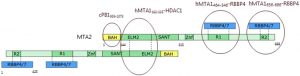
Fig.1.Diagram of the MTA2-RBBP4/7 2:4 oligomer (PMID:28179136)
Summary
Assay measuring the interaction of MTA2 to RBBP4 and RBBP7 was developed using NanoBRET technology – a proximity-based assay that can detect protein interactions by measuring energy transfer from a bioluminescent protein donor to a fluorescent protein acceptor. To determine the optimal orientation of the tags we utilized both, N- and C-terminal tagging (Fig.2.).
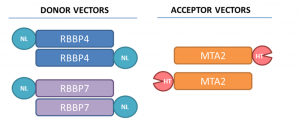
Fig.2. The tag positions in RBBP4, RBBP7, and MTA2 are indicated. HT-HaloTag, NL-NanoLuc
I determined the optimal donor to acceptor ratio of donor constructs: C- or N-terminally NanoLuc tagged RBBP4 and RBBP7 and acceptor constructs: C- or N-terminally HaloTag tagged MTA2 (Fig.3). The best results were obtained at 1:100 donor to acceptor ratio for C- terminally NanoLuc® tagged RBBP4 or RBBP7 and C- terminally HaloTag tagged MTA2 (Fig 3).
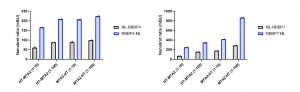
Fig 3. NanoBRET assay development for MTA2 and RBBP4 or RBBP7 interactions. U2Os cells were transfected with different donor to acceptor ratios, as indicated, and signal was read 48 h post transfection. Experimental duplicates of mean corrected NanoBRET ratios (mBU) are indicated.
We also compared the RBBP4 and RBBP7 binding to H3 at the optimized donor and acceptor plasmid ratios established above. RBBP4 and RBBP7 displayed more consistent binding to MTA2 than H3. Further experiments employing binding disrupting mutants or compounds are needed for assay validation and assay window determination.
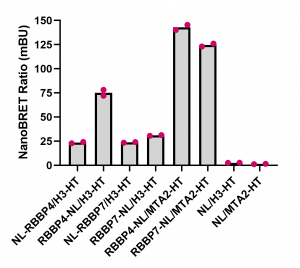
Fig. 4. Comparison of RBBP4 and RBBP7 interaction with MTA2 and H3.3. HEK293T cells were transfected with different amounts of vectors, as indicated, and signal was read 24 h post transfection. Experimental duplicates of mean corrected NanoBRET ratios (mBU) are indicated. NL denotes the NanoLuc only control, which represents background signal.
Please see the experimental details on Zenodo.
