Hello Open World!
In this very first post I will give an overview of my current and future research on malaria. I would like to sincerely thank everyone here (particularly Matthieu and Mandeep) for welcoming me so warmly.
ABSTRACT:
A major challenge for the elimination of malaria is Plasmodium falciparum asymptomatic infections, the hidden infectious reservoir. Yet I believe these infections can provide useful information on the host-pathogen interactions over an extended period of time. My research aims to understand how P. falciparum parasites can survive in chronic infections over the 6-month long dry season, using an already available collection of blood samples from a cohort of asymptomatic volunteers in The Gambia. With whole genome sequencing, we will identify parasite alleles under selection during the dry season population bottleneck. With single-cell RNA-seq, we will explore how the parasite senses its environment and adapt to it. With qPCR and flow cytometry, we will test the central dogma that antigenic variation is at the basis of a chronic infection. This work will greatly advance our understanding of the parasite, host and vector interactions, and could reveal key molecular drivers of asymptomatic infections, a critical step to enable the eradication of malaria.
INTRODUCTION:
Basic facts about malaria:
- Caused by Plasmodium parasites (unicellular eukaryote)
- Transmitted by Anopheles mosquitoes
- Plasmodium parasites spend most of their asexual life inside red blood cells, with a reinvasion cycle every 48 hours.
- Over half a million deaths per year, mainly children in Sub-Saharan Africa infected with Plasmodium falciparum
Malaria in The Gambia, West Africa
Where malaria transmission is seasonal such as The Gambia in West Africa, this occurs during and immediately after the rains (June-December). During the dry season (January-May) there is no transmission, hardly any clinical case is diagnosed during these dry months, and there are fewer mosquitoes[1,2]. Most of these infections will clear naturally, inducing a parasite population bottleneck (Fig. 1). However some P. falciparum parasites survive by establishing chronic, asymptomatic infections across the whole dry season. These infections are the reservoir from which the seasonal peak will restart at the next transmission season.
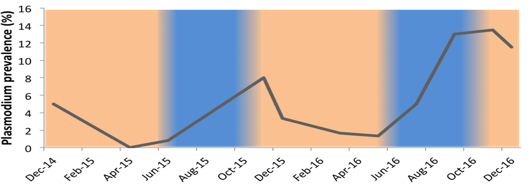 |
Figure 1. Plasmodium falciparum prevalence in a village of ~1000 inhabitants in the eastern Gambia, detected by nested PCR or quantitative PCR. The blue and orange shadings indicate the rain and dry seasons, respectively. (Claessens, unpublished data). |
My current research focuses on P. falciparum asymptomatic infections for three main reasons:
- Asymptomatic infections: out of sight, out of mind
On any given day, the vast majority of all P. falciparum infections are asymptomatic, yet they have been poorly investigated (Fig 2). Currently, our knowledge of the parasite biology is based on isolates derived from clinical cases, and usually from clonal culture-adapted parasites. How asymptomatic infections differ from clinical cases has hardly ever been investigated, with no genome, epigenome, transcriptome, proteome or phenotypic description of such parasites published to date, due to the technical challenges associated with very low amount of biological material. Today’s technology is at a turning point for addressing these issues.
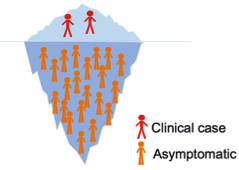 |
Figure 2. Depending on malaria endemicity, the proportion of asymptomatic infections ranges from 73 to 98% [3]. |
- Asymptomatic infections, a challenge for malaria elimination
This reservoir, shown to produce gametocytes and transmit them[4], represents the biggest challenge for malaria eradication. However, clearing all infections would include treating carriers without clinical symptoms who are unlikely to seek treatment. A campaign that would only target clinical cases, the most virulent forms of the parasite, is at risk of inadvertently selecting for a population with an “asymptomatic profile” (i.e. low-parasitaemia, chronic infections), complicating the elimination effort.
- Asymptomatic infections as a new model for host-pathogen interactions
Currently, pathogenesis studies typically collect a single blood sample from the patient on its arrival at hospital, giving us only a snapshot of the host-pathogen interaction. I believe we could “record the video” of the host-pathogen interaction with regular blood sampling of the same chronic asymptomatic carrier, to study the evolution of the same infection in real time (Fig 3).
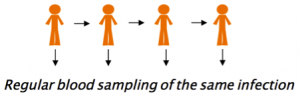 Figure 3
Figure 3
This is why in The Gambia we recruited a cohort of ~50 volunteers who were asymptomatic carriers at the start of the dry season (December 2016). These individuals were sampled monthly for up to 6 months. Epidemiology results will be described in a separate post.
Understanding how the parasite can survive in a human host for months will help us develop novel genetic surveillance & therapeutic tools.
AIMS:
To understand how the parasite is able to survive over the dry season, and start a new epidemic at the following wet season, we will characterize P. falciparum from chronic infections, at the genomic, transcriptomic and phenotypic levels, as well as the associated host immune response. More specifically:
Project 1: Identifying P. falciparum alleles under selection during the dry season
Sequencing of P. falciparum genomes collected during the wet and dry season will identify parasite alleles that are selected for by the parasite population bottleneck due to the lack of transmission in the dry season.
Project 2: Can P. falciparum sense its environment and adapt to it via transcriptional regulation?
2a: We will measure the in vitro parasite multiplication rate to determine whether isolates from asymptomatic infections grow slower than virulent infections.
2b: Using single-cell (sc) RNA-seq, we will compare the transcriptome of parasites collected in dry and wet seasons. Our hypothesis is that at the start of the dry season some subpopulations of parasites switch to a dormant-like state.
Project 3: Can antigenic variation explain chronic infections?
We will record var gene expression switching and its associated immune response monthly over the course of the same infections. This will reveal a long postulated but never formally tested hypothesis that variant surface antigen switching is sufficient for the parasite to evade the host immune system.
References:
1 Ceesay, S.J. et al. (2008) Changes in malaria indices between 1999 and 2007 in The Gambia: a retrospective analysis. Lancet 372, 1545–54
2 Ceesay, S.J. et al. (2010) Continued decline of malaria in The Gambia with implications for elimination. PLoS One 5, 4–13
3 Lindblade, K. a et al. (2013) The silent threat: asymptomatic parasitemia and malaria transmission. Expert Rev. Anti. Infect. Ther. 11, 623–39
4 Stone, W. et al. (2015) Assessing the infectious reservoir of falciparum malaria: Past and future. Trends Parasitol. 31, 287–296

This is absolutely awesome that you are doing this! Open science rules!
Very useful for our current works in mathematical models of control malaria, as we can incorporate now this annual cyclicity and consider two separate groups of infectious.
Thanks for sharing under open science.
All the best