In my last post, I showed you the genetic variants at the ADP-ribose (ADPr) binding site of SARS-CoV-2 nsp3-Mac1 and the predicted effects of those mutations on ADPr binding with Mac1.
Next, we wanted to assess the effect of all possible mutations at the sidechains lining the ADPr binding site of nsp3-Mac1 and how that would affect ADPr binding. ADPr is the endogenous ligand for nsp3-Mac1 that Michalska and their colleagues have crystalized with SARS-CoV-2 Mac1 (PDB: 6w02).
We created an energy matrix where we consider the effect of every possible mutation on the binding of ADPr with nsp3-Mac1. Table 1 includes a list of 29 residues lining the pockets.
It is likely that the residues of the rows with green color are not making optimal interactions with ADPr. This is reflected in the very slight and non-significant lowering in ddGbind values of these residues (mutations are stabilizing) and hence the green color. On the other hand, the sidechains whose mutations are shown in orange are likely the positions where macrodomain would better interact with ADPr. As a result, the mutations of those sidechains are predicted to penalize its binding significantly and hence the orange color.
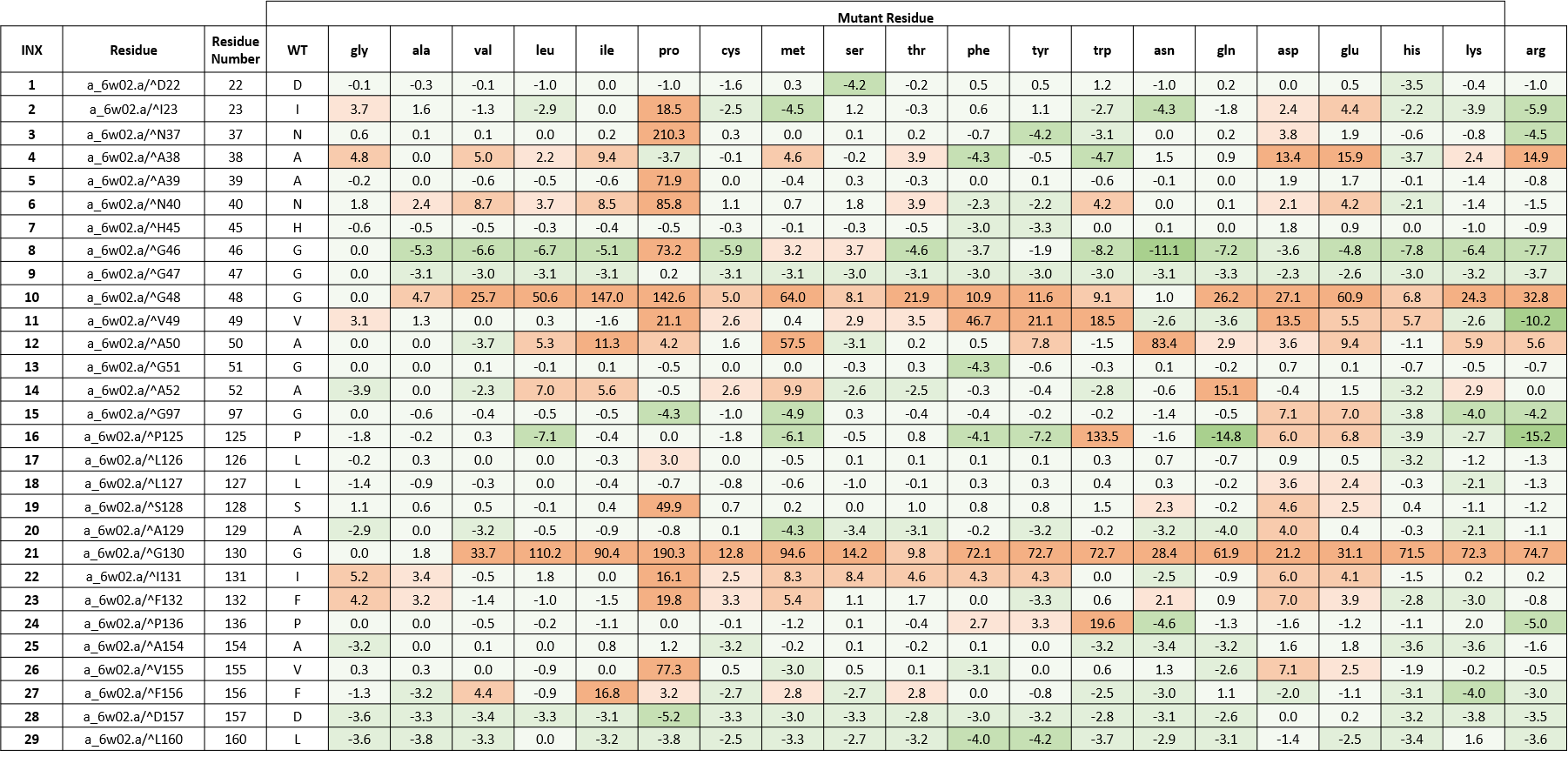
Table 1. The energy matrix showing the effect of all possible mutations at SARS-COV-2 nsp3-Mac1 ADPr binding site and the predicted effect on ADPr binding. The energy unit is kcal/mol. WT = wild type residue.
Most of the hydrogen bonds in ADPr binding pocket make use of main chain atoms and only a few hydrogen bonds involve protein side chains. Since main sidechain interactions can be achieved using several combinations of sidechains, there is less pressure on amino acid preservation. (Michalska et al., 2020)
In the absence of a catalytic residue at ADPr-binding site, Jankevicius et al. proposed substrate-assisted catalysis. Through this enzymatic mechanism, a water molecule is responsible for nucleophilic attack on the ribose anomeric carbon atom which is activated by the Pa group. (Jankevicius et al., 2013) Based on the current macrodomain/ADPr structure (PDB: 6w02), the candidate water (Wat) molecule binds to the amide group of A50, also the carbonyl of A38, and the oxygen atom of Pa and OH1’ group of the proximal ribose ring of ADPr which is shown in figure 1. (Michalska et al., 2020)
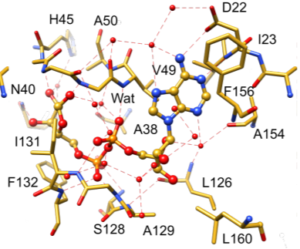
Figure 1. ADPr binding site in nsp3-Mac1 (PDB:6w02). (Michalska et al., 2020)
Based on the diversity dendrograms in post 14, it is evident that A50 is preserved among the entries of Alpha- and Betacoronavirus entries while A38 is not. On the other hand, both A38 and A50 are conserved in the SARS-CoV-2 samples from the samples of COVID-19 patients Nicola De Maio has looked at.
From table 1, we see that many cells of the rows belonging to A38 and A50 are highlighted in orange which further points at the importance of these two amino acids in substrate-assisted catalysis proposed by Jankevicius et al. (Jankevicius et al., 2013).
Conclusion
We see a lot of structural diversity at the ADP-ribose binding site of the first macrodomain of SARS-CoV-2 nsp3, both across diverse coronaviruses and across diverse SARS-CoV-2 samples. However, most of these variations are predicted to have little impact on ADPr binding. While this structural diversity is not favorable for the design of broad-spectrum ligands, competitive inhibitors mimicking interactions made by ADPr may have a broad spectrum of action.
This post marks the end of nsp3-Mac1 analysis. To see the full report on nsp3-Mac1, please refer to my Zenodo report.
Please contact me via the “Leave a comment” link at the top of this post. Stay Tuned for more updates on this project!
References:
Michalska, K., Kim, Y., Jedrzejczak, R., Maltseva, N. I., Stols, L., Endres, M., & Joachimiak, A. (2020). Crystal structures of SARS-Cov-2 ADP-ribose phosphatase (ADRP): From the apo form to ligand complexes. https://doi.org/10.1101/2020.05.14.096081
Jankevicius, G., Hassler, M., Golia, B., Rybin, V., Zacharias, M., Timinszky, G., & Ladurner, A. G. (2013). A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nature Structural & Molecular Biology, 20(4), 508-514. https://doi.org/10.1038/nsmb.2523
