Background
PRMT1 is the type I protein arginine methyltransferase (Fig.1) which mono- and asymmetrically dimethylates histone and nonhistone proteins, mainly in glycine and arginine-rich motifs and is involved in various cellular processes such as transcription, RNA processing, DNA repair, cell cycle or signal transduction (PMID:26612103). It is responsible for depositing over 90% of the total asymmetric arginine dimethyl mark (PMID:23419748). It has been identified, that at least seven distinct PRMT1 isoforms are generated by complex alternative splicing in the 5’ region of its pre-mRNA, however, their exact role has not been fully understood (PMID:11448779, 11387442). Deregulated PRMT1 expression has been observed in a number of tumor types including brain, lung, breast, colon, bladder, and leukemia, which make it a potential target for anti-cancer therapy (PMID:23235912, 28061334).
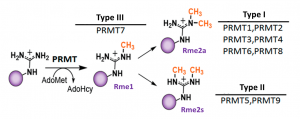
Fig.1. Mammalian PRMTs. There are 9 members and 3 types of the PRMT family. Type I, II and III PRMTs catalyze the formation of monomethyl arginines (Rme1). Type I PRMTs catalyze asymmetric arginine methylation (Rme2a), while type II PRMTs form symmetric arginine methylation (Rme2s). Type III PRMT can only monomethylate arginine residues. Modified from (PMID:29378138).
PRMT1 assay validation
It has been reported that PRMT1 is the main contributor to H4R3 (histone 4 arginine 3) asymmetric dimethylation (H4R3me2a) in cells (PMID:17848568). Moreover, with the loss of PRMT1, a large number of substrates become targets for Type II and III PRMTs, because these substrates are presumably no longer blocked by Rme2a modification (PMID: 23419748). We found that knocking down of PRMT1, but not PRMT3, 4, 5, 6, or 7 decreased basal H4R3me2a levels and increased Rme1 and Rme2s levels in MCF7 cells (Fig.2). MCF7 cell line was chosen for the PRMT1 cellular assay due to the high basal levels of H4R3me2a (Fig.3). The assay was further validated with PRMT Type I chemical probe MS023 (PMID:26598975). MS023 decreased the levels of H4R3me2a in a dose-dependent manner (Fig. 4). The Z factor for the assay equals 0.83 (n=3).
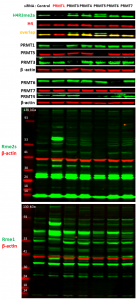
Fig.2. PRMT1 knock-down results in decrease in histone 4 arginine 3 asymmetric dimethylation levels and increase in global arginine mono- and dimethylation. MCF7 cells were treated with siRNA targeted for different PRMTs and control siRNA for 6 days and analysed by western blot for PRMTs expression, histone 4 arginine 3 asymmetric dimethylation (H4R3me2a), symmetric arginine methylation (Rme2s) and monomethylation (Rme1) levels.
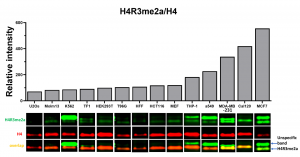
Fig.3. MCF7 cells have high basal levels of H4R3Rme2a mark. Western blot analysis of basal H4R3me2a levels in various cell lines. The bars represent H4R3me2a fluorescent intensities normalized to total H4 (n=1).

Fig.4. MS023 decreases PRMT1 dependent H4R3 asymmetric dimethylation in cells. MCF7 cells were treated with inhibitor at indicated concentrations for 48 h and H4R3me2a levels were determined by western blot. The graph represents nonlinear fits of H4R3me2a signal intensities normalized to total histone H4. The results are mean +/- SEM of 3 replicates.
For assay details, please go to Zenodo.
