Introduction to Probe my pathway (PMP): accessible at apps.thesgc.org/pmp/
Despite the sequence of the whole human genome 20 years ago, our knowledge of the human proteome remains fragmented. While ~65% of the human proteome has been investigated in varying degrees, about 35% of the human proteome remains uncharacterized (the ‘dark proteome’). A key obstacle stalling the progress in characterizing the dark proteome is the lack of tools to study protein function. In efforts to tackle this obstacle, biomedical scientists around the globe have formed a federation, called Target 2035, to develop and apply new technologies to create chemical probes for the druggable human proteome by 2035.1,2
Chemical probes are selective and potent small-molecule modulators (often an inhibitor) of proteins that can be used in biochemical, cell-based, or animal models. They are highly impactful in biomedical research, especially in understanding protein function and validating potential therapeutic targets. They have a plethora of advantages; they are complementary to genetic approaches (i.e. RNAi and CRISPR), they can be used in almost any cell type, and their actions mimic that of therapeutic small molecule drugs and thus are more relevant for translational studies. One key criteria to consider before using a chemical probe is to ensure it is of high-quality as the use of poor quality probes can generate misleading and incorrect results. The Structural Genomics Consortium (SGC) defines high-quality probes as: has in vitro potency of <100nM, has >30-fold selectivity relative to other closely-related proteins, profiled against ‘industry-standard’ selection of pharmacologically relevant off targets and protein families relevant in drug discovery, and has shown on-target effects in cells at <1µM.3
To help navigate the development of new probes and tools, we created a tool called Probe My Pathway (PMP) where I mapped expert-curated probes and chemical tools onto protein targets of human biological pathways. I did the data curation and analysis under the supervision of Dr. Matthieu Schapira and Lihua Liu helped to translate this data onto the web interface. I obtained the compounds from three different sources, Chemical Probes Portal (CPP; reviewed, in cell rating >= 3 stars), KCGS, and EUbOPEN, all of which are part of the Target 2035 initiative. The probes have been labeled with a prefix indicating the datasource they originate from: ‘CPP_’ for CPP, ‘KCGS_’ for KCGS, and ‘EUb_’ for EUbOPEN. The list of human biological pathways and their corresponding targets/Uniprot IDs was collected from Reactome: a peer-reviewed and manually curated pathway database. PMP will be updated every quarter as these data sources continue to grow with new probe, target, and pathway discoveries.
-
Icicle Charts
I’ve created icicle charts that reflect the mapping of the probes onto the Reactome pathways and the hierarchical relationship between the pathways: one for the CPP probes, one for the the kinase chemogenomics set (KCGS+EUbOPEN), and one for all compounds (CPP+kinase chemogenomics set). The pathways are shaded based on the percentage of the targets reported in Reactome for that pathway that have chemical tools. The darker the colour, the higher the percentage.
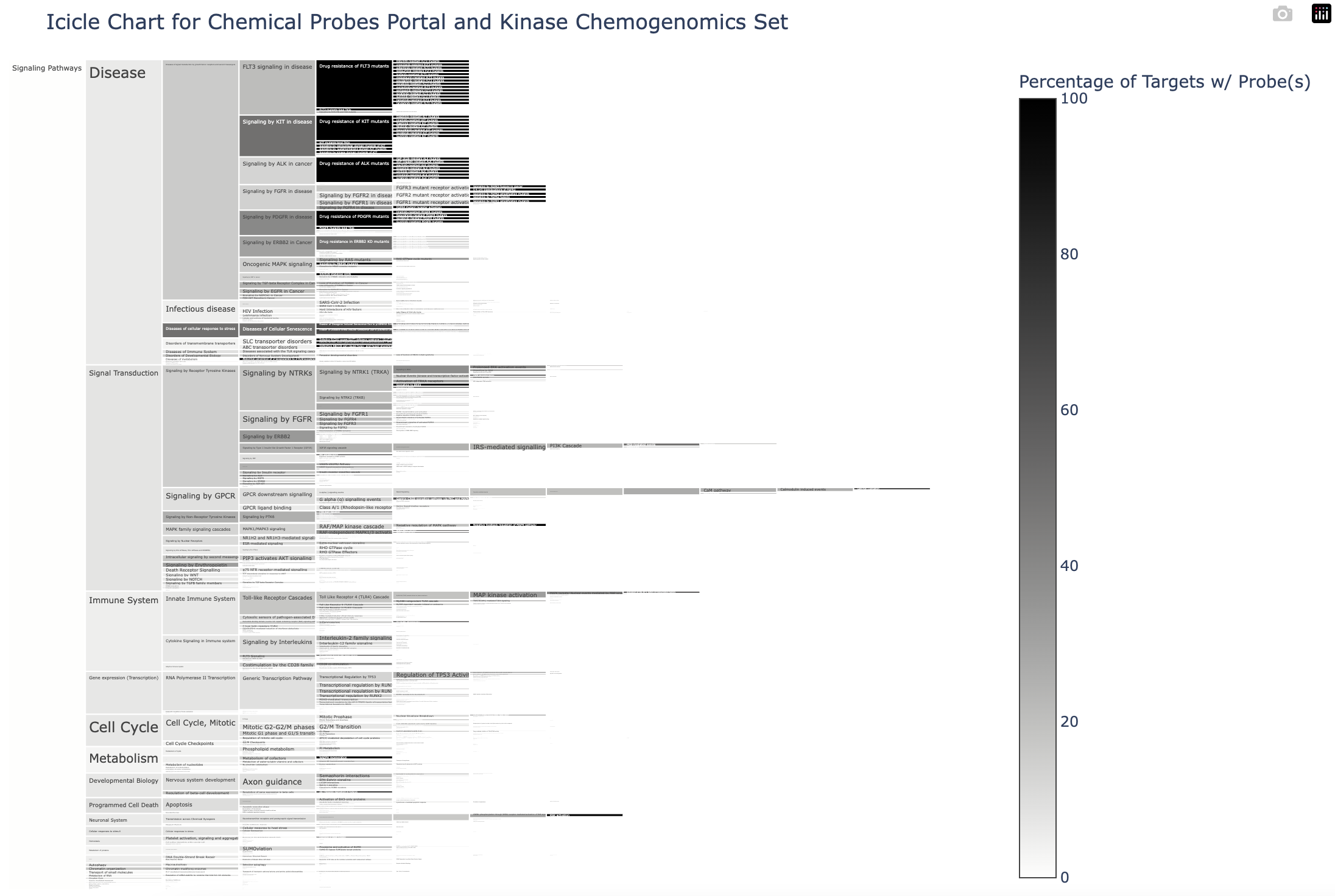
When the user hovers the mouse over a pathway, a text box appears with:
- The name of the pathway (‘Signaling by Overexpressed Wild-Type EGFR in Cancer‘)
- The Reactome pathway ID (‘Pathway ID: R-HSA-5638302‘)
- Number and percentage of targets reported in Reactome for pathway that have probe(s) (‘1/8(12%) targets have probe(s)‘)
- The name of the targets that have probe(s) and the name of the probe(s) in the format ‘Target[Probes]‘

The user can also click on the pathways to navigate to a zoomed-in view of any of the lower level (child) pathways. A bar with a list of the preceding pathways is displayed at the top of the chart; these pathways can be clicked to navigate to the previous view.

2. Search Engine
The user can search any probe, target, or pathway of their interest, and a table reflecting the data shown on the icicle charts is returned.
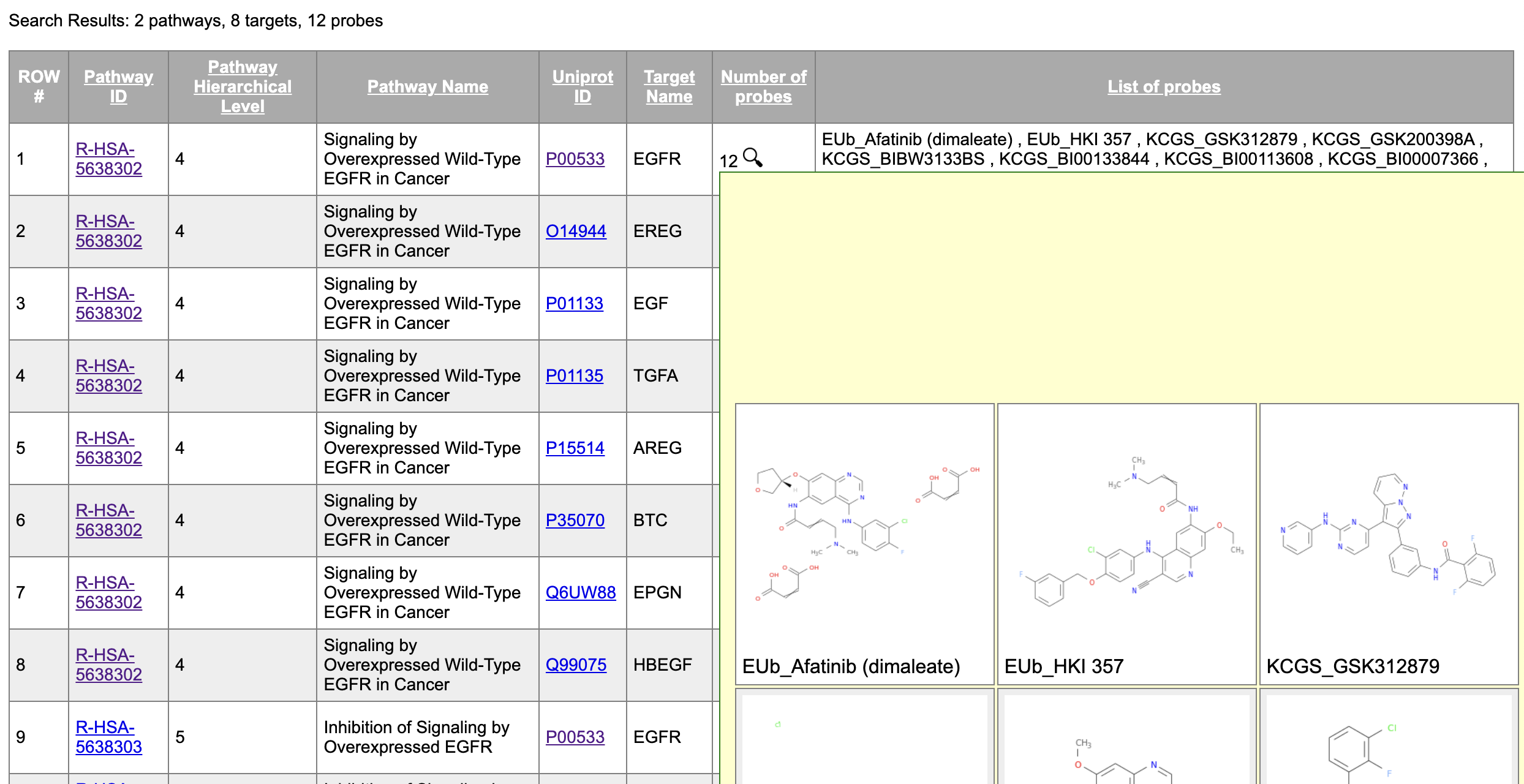
The format of the table returned is such that each row is for a single, unique protein/target. The Pathway ID is reported as a link to pathway in the Reactome website, where detailed information about the pathway can be found. The Uniprot ID is reported as a link to the information page of the protein on uniprot.org. When the user hovers their mouse over the magnifying glass icon in the ‘Number of probes’ column, the 2D structures of the corresponding probes are provided.
I will be posted about examples of possible use of PMP shortly. Stay tuned!
- Carter AJ, Kraemer O, Zwick M, Mueller-Fahrnow A, Arrowsmith CH, Edwards AM. Target 2035: probing the human proteome. Drug Discov Today. 2019;24(11):2111-2115. doi:10.1016/j.drudis.2019.06.020
- Müller S, Ackloo S, Al Chawaf A, et al. Target 2035 – update on the quest for a probe for every protein. RSC Med Chem. 2021;13(1):13-21. Published 2021 Dec 3. doi:10.1039/d1md00228g
- Arrowsmith CH, Audia JE, Austin C, et al. The promise and peril of chemical probes [published correction appears in Nat Chem Biol. 2015 Aug;11(8):541. Roth, Brian [Corrected to Roth, Bryan] and Frederiksen, Mathias [Added]] [published correction appears in Nat Chem Biol. 2015 Nov;11(11):887]. Nat Chem Biol. 2015;11(8):536-541. doi:10.1038/nchembio.1867
