One problem with working with the isolated kinase domains of ALK2 and it’s type II binding partners, is that the affinity between the kinase domains alone is insufficient to allow complex formation. We know that the two proteins associate with each other in vivo and are co-localised by the domains on the outside of the membrane. However when we just mix the two kinase domains we don’t see a strong enough association to allow a complex to be purified. We know from in-vitro tests such as the phosphorylation tests I’ve mentioned previously that there must at least be transient contact to allow phosphorylation but that isn’t sufficient to set up crystal plates of the complex.
One way to try and get around this is to artificially tether the two kinase domains together with a flexible loop. I have a construct which does exactly this, tethering one end of ACVR2 to ALK2. It’s known as QQ02ACVR2A. I purified this protein and then formed a complex with the binding partner FKBP12 which will bind to the GS domain of ALK2 present in the construct.
The first test during purification was to see if a complex could be assembled between the fusion construct and FKBP12. As they are very differences sizes (72 kDa vs. 12 kDa) purification by gel filtration gives a very clear answer. We can clearly see co-elution of the fusion and FKBP12 which is only possible if they were present as a complex. This in its self is quite exciting as it shows the fusion is still able to bind to its physiological binding partner.
The second stage after purification is to attempt to crystallise the complex. I concentrated the purified protein down and added compound K02288. K02288 definitely binds ALK2 and has some affinity for ACVR2 which is necessary for stabilising both kinase domains sufficiently to crystallise. I’ve tried crystallising ALK2 alone without compound before and had zero success so it was important to add a compound to this crystallisation attempt.
I set up four coarse screen plates and got the following in the HCS screen:
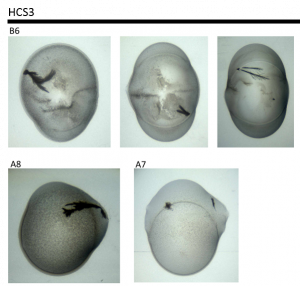
B6: 20% PEG8000, 0.2M Mg Acetate, 0.1M Cacodylate pH6.5
A7: 1.4M Na Acetate, 0.1M Cacodylate pH6.5
A8: 30% 2-propanol, 0.2M Na Citrate tribasic, 0.1M Cacodylate pH6.5
These are crystalline but not crystals so I designed a follow up screen based around these conditions. I then used what was left of my complex to set up two plates in this followup screen at 4C and 20C.
After 7 days I found the following in my plates:
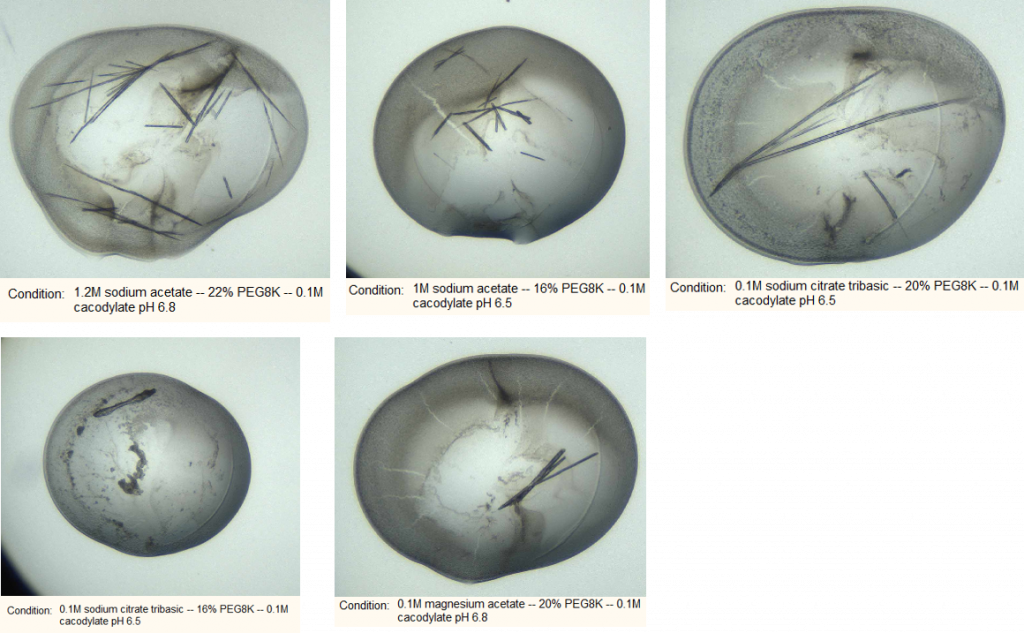
I mounted five examples and sent them to the synchrotron. Alas they all came back as either salt or no diffraction. While the salt must be ignored, it’s possible that the no diffraction crystals are protein but just do not diffract. The next step will be to make seed stocks from these wells, purify more protein and set up some more plates to see if I can get any better crystals.
You can find more details over at Zenodo.
I’m off for Christmas the middle of next week but in the new year I’ll be trying again at crystallising a variety of different proteins and compounds as well as working on characterising the phospho-mutants of ALK2 I posted about last time. Have a great holiday!
