In my previous purification of the HTT N-HEAT_81-1643 construct, I showed that an additional heparin step did not remove any traces of nucleic acid material, changed purity or oligomeric state of the sample. Additionally, my DLS results showed that this construct forms large particles in solution. Thus, the DLS results in combination with the gel filtration data gathered from the first and second rounds of purification imply that the HTT N-HEAT_81-1643 construct is in a high oligomeric state in solution. However, it is still unclear if these large particles in solution are the result of oligomerization, a monomeric state in an extended conformation (which would also register as large particles in both DLS and gel filtration chromatography), or binding to nucleic acid material. In order to learn more about the oligomeric state of this construct, I aimed at finding ideal buffer conditions to change the oligomeric state of the sample and favour the monomeric state.
First, I performed a preliminary buffer screen using DSF and DSLS. However, no differences in the temperature of aggregation (Tagg) or melting temperature (Tm) were observed in the initial conditions tested. Furthermore, the DSF data were difficult to fit due to the high initial fluorescence intensity. Like DSF, performing the buffer screen with DSLS would have been challenging to do. This is because our data showed that the Tagg for this construct lies right at the detection limit of the technique (approx. 90 oC) meaning that changes in Tagg would have been difficult to assess.
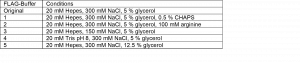
Second, I considered doing the buffer screen by changing the purification buffers and directly assessing the oligomeric state of the sample by gel filtration chromatography. This is what I did in this round of purification. The buffer screen tested the conditions listed in Table 1. I tested the addition of detergent, arginine, varying the pH, salt and glycerol. These conditions were selected based on purification strategies found in literature that were successfully applied to full length HTT or proteins in general. For instance, addition of small amounts of detergent (CHAPS) has been successfully used to purify full HTT.3, 4 Further, including arginine in the lysis buffer has been shown to stabilize the monomeric state of proteins.5, 6
Table 1. Buffer conditions tested for purification of the HTT N-HEAT_81-1643 construct
My results show that the oligomeric state of the HTT N-HEAT_81-1643 remains unchanged as evidenced by the elution volume from the gel filtration column (Figure 2). Thus, none of the conditions tested were successful at stabilizing the monomeric state of the construct. One additional strategy that I would like to test next is to co-express this construct with HAP40, as this strategy proved successful for full length HTT.2, 4 Further, in the future I will also send this sample for analysis by negative stain which can help me learn more about the oligomeric state of this construct.

Figure 1 Gel filtration chromatography profiles for the HTT N-HEAT_81-1643 construct. The concentrated elution samples of the HTT N-HEAT_81-1643 construct produced by using different purification buffers (Table 1 – FLAG buffer 1-3) were loaded on a Superose 6 10/300 GL column using the Superose 6 buffer (A-C). The absorbance of the eluted sample is monitored at 280 nm. The total volume of the column is Vc = 23.56 mL.
References
- Huang, B.; Lucas, T.; Kueppers, C.; Dong, X. M.; Krause, M.; Bepperling, A.; Buchner, J.; Voshol, H.; Weiss, A.; Gerrits, B.; Kochanek, S., Scalable Production in Human Cells and Biochemical Characterization of Full-Length Normal and Mutant Huntingtin. Plos One 2015, 10 (3).
- Guo, Q.; Huang, B.; Cheng, J. D.; Seefelder, M.; Engler, T.; Pfeifer, G.; Oeckl, P.; Otto, M.; Moser, F.; Maurer, M.; Pautsch, A.; Baumeister, W.; Fernandez-Busnadiego, R.; Kochanek, S., The cryo-electron microscopy structure of huntingtin. Nature 2018, 555 (7694), 117-+.
- Arakawa, T.; Ejima, D.; Tsumoto, K.; Obeyama, N.; Tanaka, Y.; Kita, Y.; Timasheff, S. N., Suppression of protein interactions by arginine: A proposed mechanism of the arginine effects. Biophysical Chemistry 2007, 127 (1), 1-8.
- Tischer, A.; Lilie, H.; Rudolph, R.; Lange, C., L-arginine hydrochloride increases the solubility of folded and unfolded recombinant plasminogen activator rPA. Protein science : a publication of the Protein Society 2010, 19 (9), 1783-1795.
- Harding, R. J.; Loppnau, P.; Ackloo, S.; Lemak, A.; Hutchinson, A.; Hunt, B.; Holehouse, A. S.; Ho, J. C.; Fan, L. X.; Toledo-Sherman, L.; Seitova, A.; Arrowsmith, C. H., Design and characterization of mutant and wildtype huntingtin proteins produced from a toolkit of scalable eukaryotic expression systems. Journal of Biological Chemistry 2019, 294 (17), 6986-7001.
