Sorry I’ve not written for a while. I had some annual leave to use up and then I’ve been helping out with a residential EMBO course that was being co-organised by one of the group leaders in my department. The course was an introduction to a wide variety of biochemical and biophysical techniques. I was helping out with a purification comparison between nickel resin, talon (cobalt) resin and streptactin resin and talking a bit about thermal shift assays.
Anyway, on with the science!
TNIK Selectivity:
TNIK is a protein kinase that Ros has previously talked about as being a potential off target hit for many of the M4K compounds along with DDR1. It is highly expressed in the brain which makes it of relevance to drug discovery for DIPG. Although its function is not entirely clear we know over-expression leads to cell rounding and it has been linked to some cell signalling effects in certain cancers. What makes it of most interest to us is that it seems to phosphorylate SMAD1 in an inhibitory position away from the tail where ALK2 phosphorylates SMAD1. Theoretically phosphorylation of SMAD1 by TNIK should inhibit phosphorylation elsewhere on SMAD1 by ALK2. This might make TNIK an indirect inhibitor of ALK2 function and thus we don’t want any drug we design against ALK2 to also hit TNIK.
I was working with a student, Mira Rothweiler, who purified TNIK that we then used in further experiments. Details for the purification can be found on Zenodo along with other details of the experiments done.
I have used Tm shift analysis (which is where you look at the change in melting point of a protein caused by the binding of a ligand, the tighter it binds the more it stabilises the protein and thus the larger the shift) to look at the binding of TNIK to all the M4K compounds and compared it to the shifts seen in ALK2. I plotted this data against each other with ALK2 shifts along the x-axis and the TNIK shifts along the y-axis. When compared against a 1:1 line, anything below the line is likely to be more selective for ALK2, anything above the line is likely to be selective for TNIK and anything close to the line shows minimal selectivity.
The data shows that while there are a small handful of M4K compounds that do hit TNIK, the vast majority seem to have a relatively low effect on TNIK and a high effect on ALK2 suggesting they are selective. One of the most promising M4K targets (M4K2009) gives a 14.1 °C shift for ALK2 but only 1.8 °C for TNIK. This is really good news.
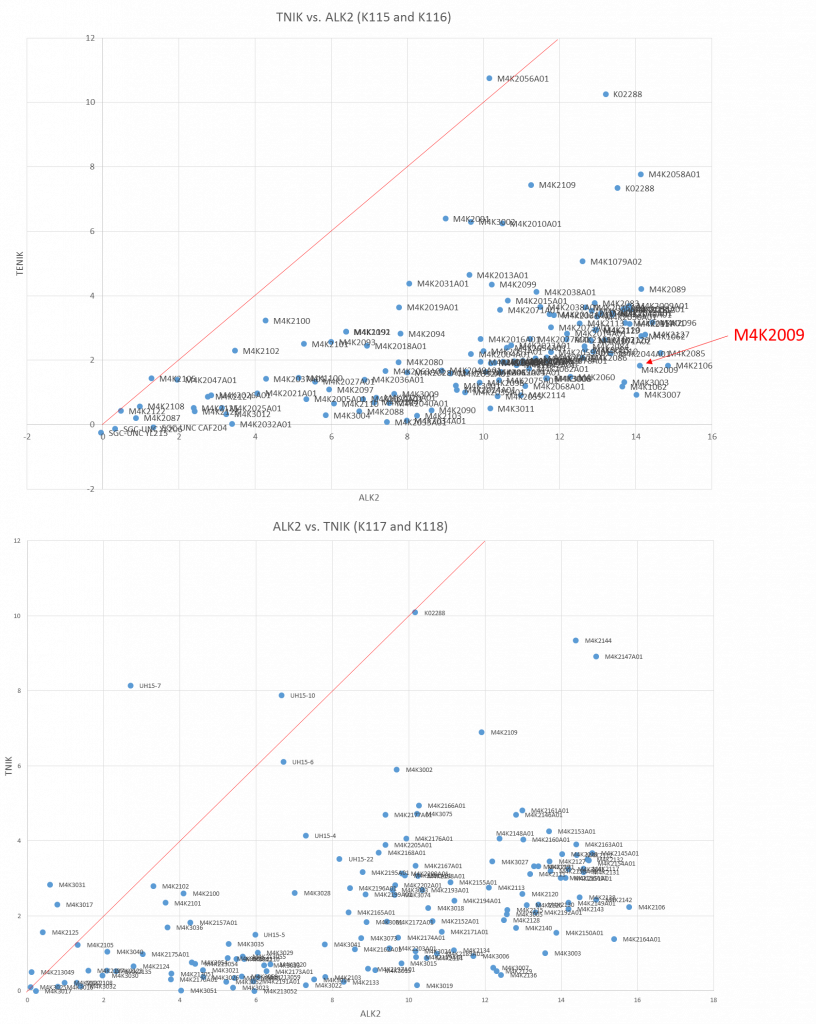
Tm shift results for ALK2 vs. TNIK for all M4K compounds. Top panel: plates K115 and K116. Bottom panel: plates K117 and K118.
TNIK activity:
While it has been reported that TNIK phosphorylates SMAD1 in a non-standard location in vivo, I wanted to see if I could get the same effect in vitro in my own experiments. I mixed SMAD1 with TNIK in different concentrations and for different lengths of time and took samples to run on mass spec to see whether SMAD1 was getting phosphorylated. My results showed that with concentrations of TNIK of 10mM and over after just 10 minutes I was starting to see phosphorylation on SMAD1. However this doesn’t tell me where that phosphorylation is happening.
I used another mass spec technique called ‘phosphomapping’ to identify where the phosphate was being added. Dr. Rod Chalk helped me with this for which I’m very thankful. This technique involved chopping the protein up into known chunks using different proteases that cut the protein at specific predictable places. We used five different proteases on different samples from the same incubation that all cut in different locations and then with each of the samples we ran half of it on the mass spec as was, and with the other half passed it over a Ti/Zr column to enrich the phosphopeptides within it and then ran it on the mass spec. The peptides detected are then run against a database of expected peptides created from knowing both the sequence of the protein and where the protease would have cut and then allowing for the possibility of modifications such as phosphorylation.
From this we detected a number of different phosphorylation sites on both SMAD1 and TNIK which are summarised in the diagram below.

Summary of identified peptides showing overall coverage (red), definitive phosphorylation (blue) and ambiguous phosphorylation (orange). Consensus site for TNIK phosphorylation of SMAD1 from Wang et. al. 2016 shown boxed in blue.
A number of sites were identified on TNIK which must be due to auto-phosphorylation. Three sites were identified on SMAD1 definitively and a further three ambiguously.
- Three of these sites (one firm and two ambiguous – PISSVS) are known sites of phosphorylation by ALK2 however we know that ALK2 cannot phosphorylate them alone and requires a type II receptor for this to happen – it seems that under in-vitro conditions TNIK is not under the same restriction.
- One of these sites (FETVY) is not known to be involved in SMAD1 inhibition but it cannot be ruled out yet and may bear further investigation
- Two of these sites (one firm and one ambiguous – RNSTIE) are in the location suggested by Wang et. al. (2016) as being the consensus sequence for TNIK phosphorylation of SMAD1.
The next thing to check will be if SMAD phosphorylated in the ‘inhibitory’ position stops ALK2 from phosphorylating SMAD in the ‘normal’ position.
You can find more detail on experimental work over on Zenodo.
Reference:
Identification of Phosphorylation Consensus Sequences and Endogenous Neuronal Substrates of the Psychiatric Risk Kinase TNIK. Wang Q, Amato SP, Rubitski DM, Hayward MM, Kormos BL, Verhoest PR, Xu L, Brandon NJ, Ehlers MD. Pharmacol Exp Ther. 2016 Feb;356(2):410-23. doi: 10.1124/jpet.115.229880. Epub 2015 Dec 8.
