My name is Joseph Newman and I have been working in Lab of Opher Gileadi at SGC Oxford since 2013 mainly on targeting DNA repair factors as synthetic lethal targets for early stage development of new cancer therapeutics. Like many scientists my normal laboratory based research activities were disrupted by the lockdown, and when Opher suggested to work on targeting the SARS-Cov-2 helicase (a family that I have significant experience and interest in) I was keen to give it a try. The SARA-Cov-2 helicase is well conserved across and appears to be indispensable across all coronavirus species, and for this reason has been highlighted as one of the attractive targets for the development of anti-viral drugs(1, 2). The helicase is a Upf1 like SF1b helicase that unwinds both DNA and RNA in a 5’ to 3’ direction with a relaxed nucleotide specificity in vitro. Whilst its precise role in viral replication in not currently well-defined recent studies show it acts in concert with the replication-transcription complex (Nsp7/Nsp82/Nsp12), possibly being involved in either disrupting downstream RNA secondary structures or template switching(3).
Previously crystal structures of Nsp13 have been solved for MERS-CoV(4) and SARS-Cov-1(5) to 3.0 Å and 2.8 Å respectively which indicated that the protein may be crystallizable, although the resolution of these structures is probably too low to reliably detect fragment binders in a X-ray Fragment screen. We began lab work on this project in mid-April and made quite rapid progress obtaining a new crystal form that is reproducible and diffracts routinely to around 2.0 Å. We have deposited the Nnsp13 structure in the pdb (6ZSL) and recently ran a full fragment screen with the help of XChem team at Diamond Light Source. The screen went about as well as could be expected producing around 50-60 hits from a screen of around 600 compounds with several compounds binding in both the nucleotide and RNA binding sites. I am still working on finalising the hits and the results should be available soon on the Diamond fragalysis user interface, but I thought I would show you an overview of what we found in the image below
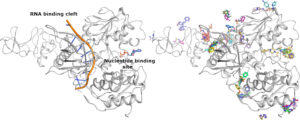
The structure of Nsp13 is shown on the left panel along with the expected positions of the Nucleotide and RNA which have been positioned into the enzyme based on structures of homologues. The right hand panel shows a subset of the fragment hits which can be seen to cluster around areas of potential interest for inhibition.
This project was a team effort and I want to thank Yuliana Yosaatmadja for help with the protein work and the whole XChem team at Diamond in particular Alice Douangamath for help in setting crystallization experiments and mounting all the crystals. If you are reading this and are interested in collaborating towards the development of Nsp13 inhibitors, then please get in touch.
For a detailed protocol of Nsp13 purification and crystallization please refer to my Zenodo post.
References
- Habtemariam S, Nabavi SF, Banach M, Berindan-Neagoe I, Sarkar K, Sil PC, et al. Should We Try SARS-CoV-2 Helicase Inhibitors for COVID-19 Therapy? Arch Med Res. 2020.
- Keum YS, Jeong YJ. Development of chemical inhibitors of the SARS coronavirus: viral helicase as a potential target. Biochem Pharmacol. 2012;84(10):1351-8.
- Chen J, Malone B, Llewellyn E, Grasso M, Shelton PMM, Olinares PDB, et al. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. bioRxiv. 2020.
- Hao W, Wojdyla JA, Zhao R, Han R, Das R, Zlatev I, et al. Crystal structure of Middle East respiratory syndrome coronavirus helicase. PLoS Pathog. 2017;13(6):e1006474.
- Jia Z, Yan L, Ren Z, Wu L, Wang J, Guo J, et al. Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47(12):6538-50.
