Hello!
My name is Nergis Imprachim and I am working at CMD, University of Oxford. I am working on drug discovery projects– one of them focuses on targeting T-box transcription factor T (TBXT) that is associated with chordoma. Chordoma is a bone cancer affecting the skull base and spinal cord. It is a slow-growing, rare disease with an incidence of one per one million people annually. It is poorly-responsive to radiotherapy and chemotherapy. Chordoma tumors are currently treated by surgical resection, but they recur in more than half of the cases. Chordoma patients still need more effective treatments, and as a scientist, this has brought me into action to help develop new drugs and help the patients. I am currently dedicated to a super collaborative project, supported by the Chordoma Foundation and the Mark Foundation for Cancer Research, with the purpose of fighting against chordoma.
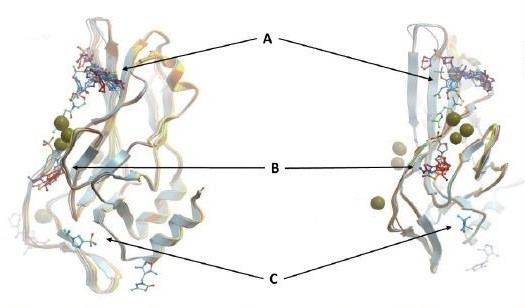
Figure 1: Overlay of some fragments with DNA-free WT TBXT DBD. Pocket A, B, and C are just some of the hot spots for TBXT inhibition
Chordomas are characterized by the expression of TBXT (the homolog of the mouse Brachyury). TBXT is expressed early during vertebrate development to help form embryonic tissues, mesoderm and notochord. It is normally turned off after day-13 in human development [3]. In chordomas, however, its expression is aberrantly maintained. In most of the chordomas, TBXT harbors the nonsynonymous single-nucleotide polymorphism (SNP) G177D in its DNA-binding domain (TBXT DBD) [1, 2]. Role of this SNP in chordoma is not yet fully understood. It is possible that G177D mutation affects binding of TBXT to DNA and/or formation of TBXT dimer, which is the more stable and functional form of the protein. Since TBXT is uniquely expressed in chordoma cells, not expressed in any post-embryonic tissues of healthy individuals, and accompanied by this functionally important SNP, we strongly believe that TBXT is an ideal therapeutic target for chordoma treatment.
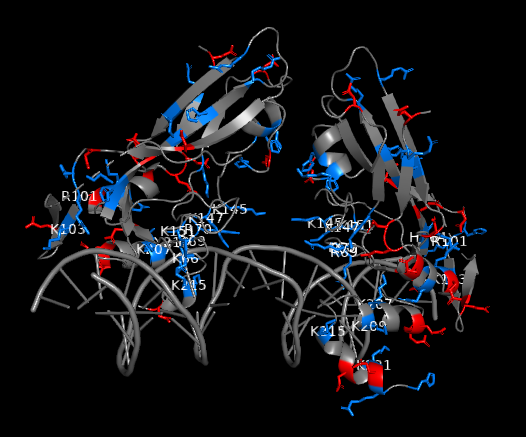
Figure 2: Charged residues of TBXT G177D mutant in complex with DNA (blue: positively charged, red: negatively charged). Positively charged residues are clustered near DNA interface. Strong polar interactions exist between the polar residues of TBXT DBD and DNA
As this potential drug target needs to be further investigated through a drug discovery research, our team set off by first solving the crystal structures of wild-type and G177D variant of TBXT DBD in complex with DNA. The team at Diamond Light Source performed crystal-based fragment screening (XChem), which allowed to screen hundreds of fragments in a short period of time. Screening results showed that there are >40 fragments bound to several different pockets within DBD. Those pockets engage with important functions of the protein, and this makes them attractive sites of inhibition. For example, one of the pockets called Pocket A is close to TBXT dimerization interface, which forms upon DNA binding (Figure 1). Dimerization interface area is small (2.3% of monomer’s accessible surface area) and interaction seems to be weak (indirect interactions through water molecules in solution). Targeting this weak interface could disrupt TBXT-DNA binding. That’s one way to inhibit TBXT. Another pocket called Pocket B is close to DNA interface and also one of the Pocket C fragments engages DNA interface. Direct targeting of DNA binding site of TBXT could be another way to inhibit the protein although it is relatively difficult to interfere with the polar interactions that we have seen between TBXT DBD and DNA (Figure 2). These are some of the sites (and there are more!) that we are currently targeting with the small molecules. You can refer to the post of our collaborator David Drewry to learn some more, there you go. And here’s another one!
We are a huge, multidisciplinary team aiming at drugging the TBXT protein. The Drewry group at the University of North Carolina at Chapel Hill work on the optimization of the initial XChem fragments into small molecules. Paul Workman and his group at the Institute of Cancer Research London work on the molecular pharmacology aspect and study how the potent small molecules behave in chordoma cells. Here at the University of Oxford, we are using biophysical (differential scanning fluorimetry (DSF, Tm shift), surface plasmon resonance (SPR), fluorescence anisotropy/polarization (FP)) and structural (X-ray crystallography) methods to investigate the binding between the small molecules and TBXT. If you have any questions about our research, please fire away! Lastly, if you are interested, here’s the experimental protocol for the expression and purification of TBXT (DNA-binding domain and full-lenght). For detailed methods, please refer to my Zenodo records (DOI: 10.5281/zenodo.5510378 and DOI: 10.5281/zenodo.5515635)
References:
[1] Pillay N, Plagnol V, Tarpey PS, Lobo SB, Presneau N, Szuhai K et al. A common single-nucleotide variant in T is strongly associated with chordoma. Nature Genetics. 2012;44: 1185–1187. https://doi.org/10.1038/ng.2419
[2] Papapetrou C, Edwards YH, Sowden JC. The T transcription factor functions as a dimer and exhibits a common human polymorphism Gly-177-Asp in the conserved DNA-binding domain. FEBS Lett. 1997;409(2): 201-6. https://doi.org/10.1016/s0014-5793(97)00506-1
[3] Sheppard HE, Dall’Agnese A, Park WD, Shamim H, Dubrulle J, Johnson HL et al. Targeted brachyury degradation disrupts a highly specific autoregulatory program controlling chordoma cell identity. Cell Reports Medicine. 2021;2(1): 100188. https://doi.org/10.1016/j.xcrm.2020.100188

Hello,
Great work! Where could one acquire such an E coli cell line (BL21 DE3 RR pRare)?
Hi there,
We generate BL21(DE3)-RR-pRARE competent cells in Oxford. We can send you some if you can pay for shipping. Those cells have some additional phage resistance, they’re not so different from other commercial BL21(DE3) Competent E. coli. Please get in touch if you’d like some.
Cheers,
Nergis