In a previous post, I tested hit analogues and found compound XSR00035795a (Figure 1) had increased potency and ligand efficiency; however, compound XSR00035795a was also found to bind to HDAC6 Zf-UBD, so it was not selective towards USP5.
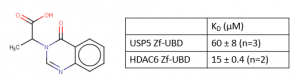 Figure 1. XSR00035795a
Figure 1. XSR00035795a
We hypothesized that it was possible to extend the aliphatic group on the carboxylic chain to further increase potency and potentially improve selectivity towards USP5 Zf-UBD. We ordered and tested these compounds- experimental details can be found on Zenodo.
Unfortunately, the follow-up compounds with extended aliphatic groups on the carboxylic chain showed no significant binding (KD >1 mM) to USP5; the extension of the methyl group does not seem to be tolerated (Figure 2).
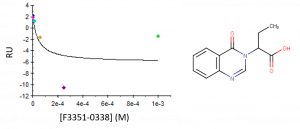
Figure 2. Binding curve of compound F3341-0338 (no binding)
Next, we’ll be looking for different compounds scaffolds through a larger virtual screen using Molecular Forecaster’s FITTED docking platform.
On another note, a chemical functionality feature is being introduced to the open lab notebook platform. This feature will allow users to upload SDF files or draw chemical structures and readers will be able to make substructure searches against all future notebook records (Figure 3).

Figure 3. Substructure Search function on the openlabnotebooks.org website
I’ve uploaded an SDF file of the compounds I tested in this experiment. A few of the compounds and pertinent information (SMILES and molecular weight) can be seen below.
| Structure Image | Smiles | Mol. Weight |
|---|---|---|
| CC(C(=O)O)n1c(=S)[nH]c2ccccc2c1=O | 250.3 | |
| C=C1Nc2ccccc2C(=O)N1C(C)C(=O)O | 232.2 | |
| CC(C(=O)O)n1cnc2ccccc2c1=O | 218.2 | |
| CC(C)C(C(=O)O)n1cnc2ccccc2c1=O | 246.3 |
