The experimental set-up and results covered in this post can be found here: 10.5281/zenodo.1292884.
A few posts back, I tried expressing HTT with different lengths of Q repeats in mammalian cells using baculovirus (read more here). However, expression was not very good, especially for HTT with longer Q repeats (have a look at these results here). Anyway, forging ahead, I decided to try the baculovirus overexpression in brain cells, which would be more relevant to HD.
As our lab generally works on cancer, we have the brain cancer lines LN18 and T98G. These lines were both established from patients with glioblastoma. Although, being brain cancer lines, these may not be ideal for the investigation of HD, especially with previous reports indicating that people with HD might have a lower incidence of cancer. Here is a study showing that the lower incidence of cancer in HD patients might be due to the mutant HTT protein causing more apoptosis in pre-cancer cells:
Sørensen SA, Fenger K, Olsen JH.
Significantly lower incidence of cancer among patients with Huntington disease: An apoptotic effect of an expanded polyglutamine tract?
Cancer. 1999 Oct 1;86(7):1342-6.
https://doi.org/10.1002/(SICI)1097-0142(19991001)86:7<1342::AID-CNCR33>3.0.CO;2-3
More recently, another paper has shown that the siRNAs based on the Q repeats in HTT could kill cancer cells!
Murmann AE, Gao QQ, Putzbach WE, Patel M, Bartom ET, Law CY, Bridgeman B, Chen S, McMahon KM, Thaxton CS, Peter ME.
Small interfering RNAs based on huntingtin trinucleotide repeats are highly toxic to cancer cells.
EMBO Rep. 2018 Mar;19(3). pii: e45336. doi: 10.15252/embr.201745336. Epub 2018 Feb 12.
http://doi.org/10.15252/embr.201745336
There is clearly a link between cancer and the etiology of HD, mutant HTT or even wild-type HTT that would be interesting to look at. So, I tried overexpressing the HTT constructs in the brain cancer cell lines LN18 and T98G. Here are my Western blot results showing levels of HTT and FLAG.
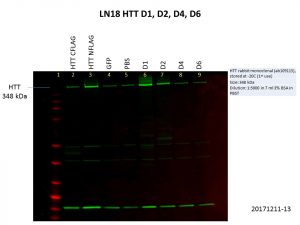
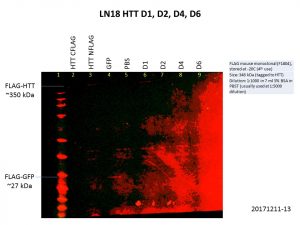
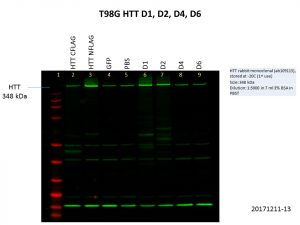
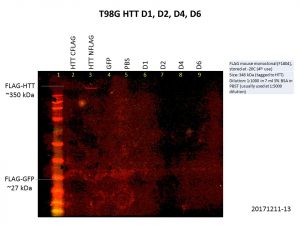
Alas, the overexpression was even worse than what I got previously! This is not surprising though, as the cells used previously, HEK293T, are known for being easy to grow and manipulate. Although there is a bit of increase in the levels of HTT, FLAG levels were barely detectable, and there was no increase in HTT levels for the D4 and D6 constructs. We might need to go back to troubleshooting ways to improve the expression of these constructs with baculovirus constructs, especially in more relevant cell lines.
