The experimental set-up and results covered in this post can be found here: 10.5281/zenodo.1323902
As I’ve been having problems overexpression HTT properly in my cancer cell lines, I decided to continue looking at other proteins of interest using our best overexpression model so far, which is in HEK293T cells. Although it may not be a cancer cell line, it is very widely used due to the ease of genetic manipulation, as evident in my success with overexpressing HTT in this line, and would be useful for the elucidation of possible mechanisms of action, which can then be validated in other more relevant but difficult cells.
On my hunt for possible target proteins to look at in my HTT overexpression model, I found several interesting papers on posttranslational modifications of proteins. Posttranslational modifications occur on proteins after they have been translated from RNA to protein and are important in the normal function of proteins, including ensuring that proteins are folded properly and do not aggregate. As the pathological feature of Huntington’s disease is the formation of protein aggregates due to the expanded polyQ region, it is highly relevant to look at changes in posttranslational modifications in the context of huntingtin overexpression.
PRMT5- mediated symmetric arginine dimethylation is attenuated by mutant huntingtin and is impaired in Huntington’s disease (HD)
Ratovitski T, Arbez N, Stewart JC, Chighladze E, Ross CA.
Cell Cycle. 2015;14(11):1716-29.
https://doi.org/10.1080/15384101.2015.1033595
This paper showed that arginine dimethylation was decreased by mutant huntingtin in Huntington’s disease. Interestingly, normal HTT, but not mutant HTT, stimulated arginine methylation mediated by PRMT5 activity. The main epigenetic mark mediated by PRMT5 is symmetric dimethylation of arginine (RMe2s). I thus decided to look at the levels of RMe2s in cells with HTT OE to see if there were any changes in PRMT5 activity. As a control, I also looked at asymmetric demethylation of arginine (RMe2a), which should not be affected by changes in PRMT5 activity.
I also found several other pathways that have been reported to be disrupted by HTT. In particular, HTT is known to be important for nuclear integrity and nuclear export, and several papers looked at how the nuclear lamina is regulated by Huntingtin. I decided to examine the nuclear integrity of HTT OE cells by looking at Lamin B1, which is a protein involved in the nuclear lamina and which was also used in some of these papers to study nuclear integrity.
Polyglutamine-Expanded Huntingtin Exacerbates Age-Related Disruption of Nuclear Integrity and Nucleocytoplasmic Transport
Gasset-Rosa F, Chillon-Marinas C, Goginashvili A, Atwal RS, Artates JW, Tabet R, Wheeler VC, Bang AG, Cleveland DW, Lagier-Tourenne C.
Neuron. 2017 Apr 5;94(1):48-57.e4.
https://doi.org/10.1016/j.neuron.2017.03.027
Focal distortion of the nuclear envelope by huntingtin aggregates revealed by lamin immunostaining
Chapple JP, Bros-Facer V, Butler R, Gallo JM.
Neurosci Lett. 2008 Dec 12;447(2-3):172-4. Epub 2008 Oct 1.
https://doi.org/10.1016/j.neulet.2008.09.075
B10 Nuclear Lamina Is Differentially Altered In Huntington’s Disease Brain Regions
J Neurol Neurosurg Psychiatry 2014;85:A12.
Alcalá R, Creus-Muncunill J, Azkona G, Alberch J, Pérez-Navarro E.
http://doi.org/10.1136/jnnp-2014-309032.38
Huntingtin was also previously reported to be involved in perinuclear localisation and centrosomal organisation. PCNT is a protein that localises to the centrosome and is involved in the formation of the pericentriolar matrix (PCM), which is key for proper centrosome formation and cell cycle progression. Its role in both perinuclear localisation and centrosomal organisation makes it an important candidate to look at in the context of Huntington’s disease. Moreover, it is also involved in ciliogenesis, which has been reported to be disrupted in patients with Huntington’s disease.
Perinuclear localization of huntingtin as a consequence of its binding to microtubules through an interaction with beta-tubulin: relevance to Huntington’s disease
Hoffner G, Kahlem P, Djian P.
J Cell Sci. 2002 Mar 1;115(Pt 5):941-8.
http://jcs.biologists.org/content/115/5/941.long
Centrosome disorganization in fibroblast cultures derived from R6/2 Huntington’s disease (HD) transgenic mice and HD patients
Sathasivam K, Woodman B, Mahal A, Bertaux F, Wanker EE, Shima DT, Bates GP.
Hum Mol Genet. 2001 Oct 1;10(21):2425-35.
http://doi.org/10.1093/hmg/10.21.2425
Ciliogenesis is regulated by a huntingtin-HAP1-PCM1 pathway and is altered in Huntington disease
Keryer G, Pineda JR, Liot G, Kim J, Dietrich P, Benstaali C, Smith K, Cordelières FP, Spassky N, Ferrante RJ, Dragatsis I, Saudou F.
J Clin Invest. 2011 Nov;121(11):4372-82. Epub 2011 Oct 10.
http://doi.org/10.1172/JCI57552
Finally, I also looked at coilin, a major component of cajal bodies. There have been reports that cajal bodies may play a role in the pathogenesis of CAG repeat diseases such as Huntington’s disease. Furthermore, it is a known substrate of PRMT5 and may be affected indirectly by Huntingtin through methylation by PRMT5.
Interaction between neuronal intranuclear inclusions and promyelocytic leukemia protein nuclear and coiled bodies in CAG repeat diseases
Yamada M, Sato T, Shimohata T, Hayashi S, Igarashi S, Tsuji S, Takahashi H.
Am J Pathol. 2001 Nov;159(5):1785-95.
http://doi.org/10.1016/S0002-9440(10)63025-8
My final list of proteins to look thus consisted of: RMe2s, RMe2a, Lamin B1, PCNT and coilin, with GAPDH as the loading control.
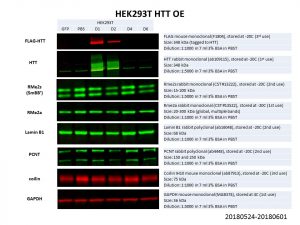
Unfortunately, none of these proteins showed any significant change in protein levels! It is possible that the HTT overexpression was not good enough, especially for the larger constructs of D4 and D6. However, even comparing between the HTT WT overexpression (D1: Q23) and the PBS control, there is no obvious difference in protein levels. Another possibility is that HTT levels is not limiting in the pathways looked at and hence an overexpression does not affect these proteins of interest. It might thus be more prudent to look at knockdowns of HTT instead.

I wonder if you might have better luck designing the experiment differently. Rather then chase down the observations of others (most of which will be artifactual), why not design the experiments in a more unbiased way?
I recall the quote “the number of proteins with which a protein interacts is directly proportional to the number of people that work on that protein”
You have more purified protein than anyone else – have you access to a kick-ass monoclonal(s)? make some?
Have the ones you used been rigorously characterized on cells and k/o cells in parallel to make sure they are specific and selective?
Hey Al, thanks for your comment! I totally agree with your quote that we tend to see interactions the more we study a particular protein or gene. I don’t think it’s an issue with the antibodies though, I get pretty clean bands for most of the targets I looked at, and the HTT antibody, which is the most important antibody since it’s for my direct protein of interest, works great (it is a rabbit monoclonal from abcam and I highly recommend it to anyone looking at HTT: https://www.abcam.com/huntingtin-antibody-epr5526-ab109115.html). However, you’re right that the experimental design can be further improved. Most of these papers that I’ve been looking at are in different lines and they have mostly been focused on mutant HTT, which I’ve had trouble overexpressing since it’s so large. Moving forward, I’ll be focusing on WT HTT for now, since the overexpression for those seem to be working, rather than trying to tease out if my experiment didn’t work because of the poor overexpression of mutant HTT.