In my previous post, I described four promising fragments bound to HAO1 structures.
Specifically, I described two key areas of HAO1 – the gating loop and active site Trp110 – where fragments 1, 2 and 5 bind. I also described fragment 6 at the subunit interface of HAO1, away from the active site.
After analysis of the fragment-bound structures, the next step is to determine how these four fragments affect the behaviour of HAO1 in solution. In this post, I will describe two biophysical assays: first, I tested whether binding of fragments could be detected in solution using surface plasmon resonance (SPR); and second, I tested whether the fragments were able to inhibit the activity of HAO1, using a fluorescence-based assay. You can find more details on Zenodo here.
Surface plasmon resonance confirmed binding of all four fragments to HAO1
To confirm that the fragments are binders of HAO1 in solution, and not merely an artefactual crystal soaking observation, I used SPR. In SPR, the protein is immobilised onto a planar metal surface and a beam of polarised light is bounced off the surface. A solution containing the molecule of interest (in this case, varying concentrations of each fragment) is passed over the immobilised protein. When a fragment binds, this changes the intensity of light reflected at different angles – the amount this changes (the ‘response’) is directly proportional to the strength of binding (figure 1A).
Using SPR, we were able to confirm that all four fragments bind to HAO1 at fragment concentrations from 15 to 100 µM (figure 1B). However, due to the relatively small sizes of fragments, they bind too weakly for accurate dissociation constants (Kd) to be derived from a plot of response vs. fragment concentration.
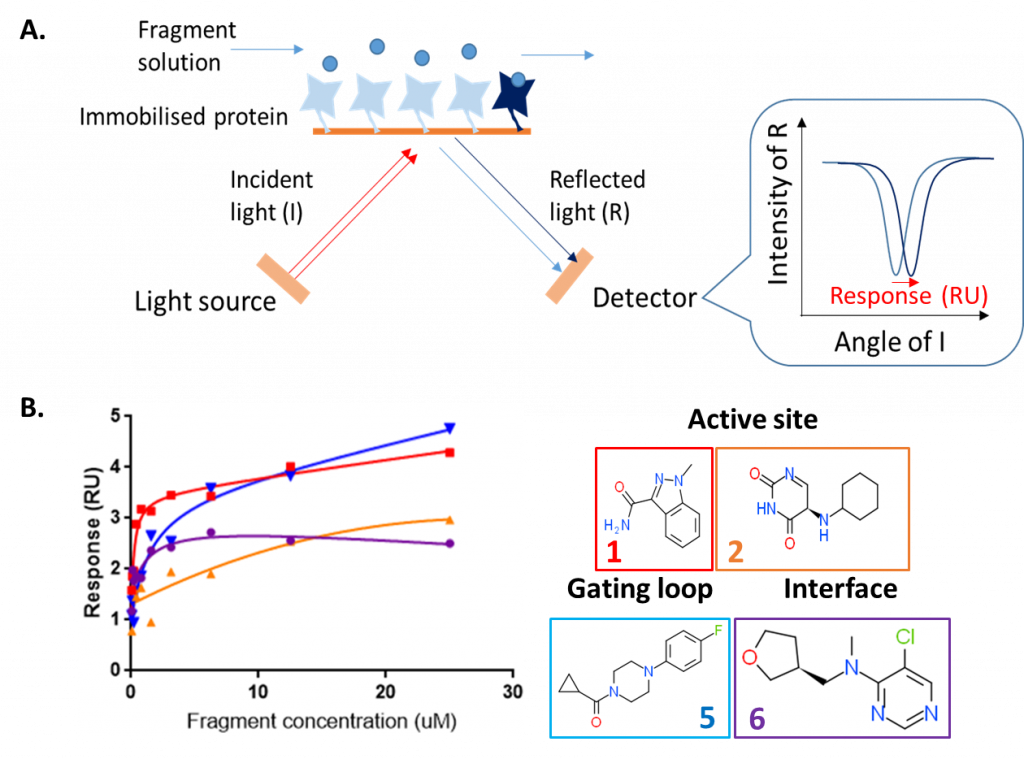
Figure 1: Surface plasmon resonance assay to detect binding of HAO1 fragment hits in solution. A. Principle of surface plasmon resonance B. Results for four selected fragments.
Fluorescence-based coupled enzyme assay confirmed inhibition of HAO1 by fragments 1, 2 and 5
To measure HAO1 enzyme activity, I used the production of hydrogen peroxide (H2O2) from molecular oxygen by HAO1 when it converts glycolate to glyoxylate. H2O2 production can be monitored by coupling to hydrogen peroxidase, which uses H2O2 to convert a colourless chemical, Amplex Red, to a fluorescent dye, resorufin (figure 2A). The level of fluorescence is directly proportional to the amount of H2O2 – and so the amount of glyoxylate – produced, allowing us to compare how active HAO1 is in the presence or absence of the fragments.
Using this coupled assay, I confirmed that the two active site fragments and the gating loop fragment inhibit HAO1 activity (figure 2B). As expected, the two active site fragments were the most potent, inhibiting up to 85% (fragment 1) and 37% (fragment 2) of HAO1 activity at the maximum tested concentration (10 mM fragment). The gating loop fragment (fragment 5) also showed detectable inhibition of HAO1 (19%) at this concentration. No inhibition was observed with the interface fragment (fragment 6), which makes sense since the fragment is too distant from the active site for direct competitive inhibition, yet is likely too small to disrupt the tetramer and HAO1 activity indirectly.
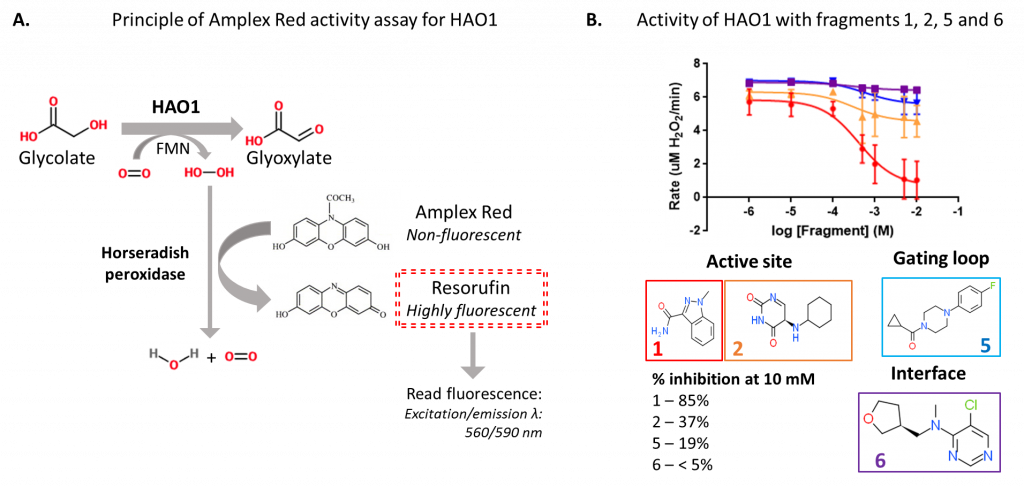
Figure 2: Fluorescence-based activity assay to detect inhibition of HAO1 by fragment hits. A. Principle of Amplex Red assay. B. Results for four selected fragments.
In my next post I will discuss how I am optimising the active site fragment hits to generate more potent inhibitors of HAO1.

One Reply to “Characterisation of HAO1 fragment hits in solution”