Now that I’ve got my biophysical screening assays developed (19F NMR, SPR), as well as my hit expansion campaign started for small molecule inhibitors against the USP5 Zf-UBD, I need to be able to test if these small molecule inhibitors will antagonize the catalytic activity of USP5.
It has been speculated in literature that the Zf-UBD is responsible for substrate recruitment, substrate positioning for proper conformation for cleavage, and that binding of ubiquitin at the Zf-UBD allosterically activates catalytic cleavage; however, the exact function of the Zf-UBD is still unclear. Perhaps, the Zf-UBD is more important for the cleavage of some substrates than others. For this reason, I need an assay that will work with diverse substrates to elucidate the function of the Zf-UBD in regards to catalytic activity.
I was able to express and purify full length USP5 and SGC’s Dr. Suzanne Ackloo developed and optimized a mass spectrometry (MS) assay to detect polyubiquitin cleavage. You can see full experimental details on Zenodo. Mass spectrometry is an analytical tool used to sort ions on the basis of their mass-charge ratio; this allows for the measurements of masses within a sample. Before the reaction begins I am able to see a clear intensity of the substrate, a tetra-ubiquitin species (Ub4K63). After the addition of USP5, over time the tetra-ubiquitin (Ub4) is cleaved into tri, di and mono-ubiquitin (Ub3, Ub2, Ub1) (Figure 1).
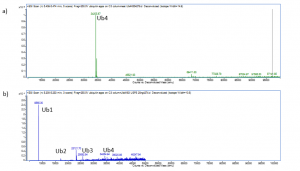
Figure 1. Mass spectra of a) Substrate Ub4K63 b) 20 ng USP5 reaction at 1 hour
Next, I’ll be testing some of the small molecule inhibitors of the zinc-finger ubiquitin binding domain (Zf-UBD) in the mass spectrometry assay to determine if the small molecules can antagonize the catalytic activity of USP5.
