Further down the line I want to be able to test the activation of the BMP signalling pathway downstream of the ALK2 receptor. For this I plan to use the alkaline phosphatase gene as a readout – it’s a well known gene activated downstream of BMP signalling so a lot of easy to use assays already exist for it.
Unfortunately when I tested this assay with the standard C2C12 cell line it didn’t work particularly well so I’ve decided to try again with a new set of cells. This time I’m choosing connective tissue cells (fibroblasts) from FOP patients as a reporter cell line because they respond more strongly to the signals that activate the BMP pathway (called BMP ligands).
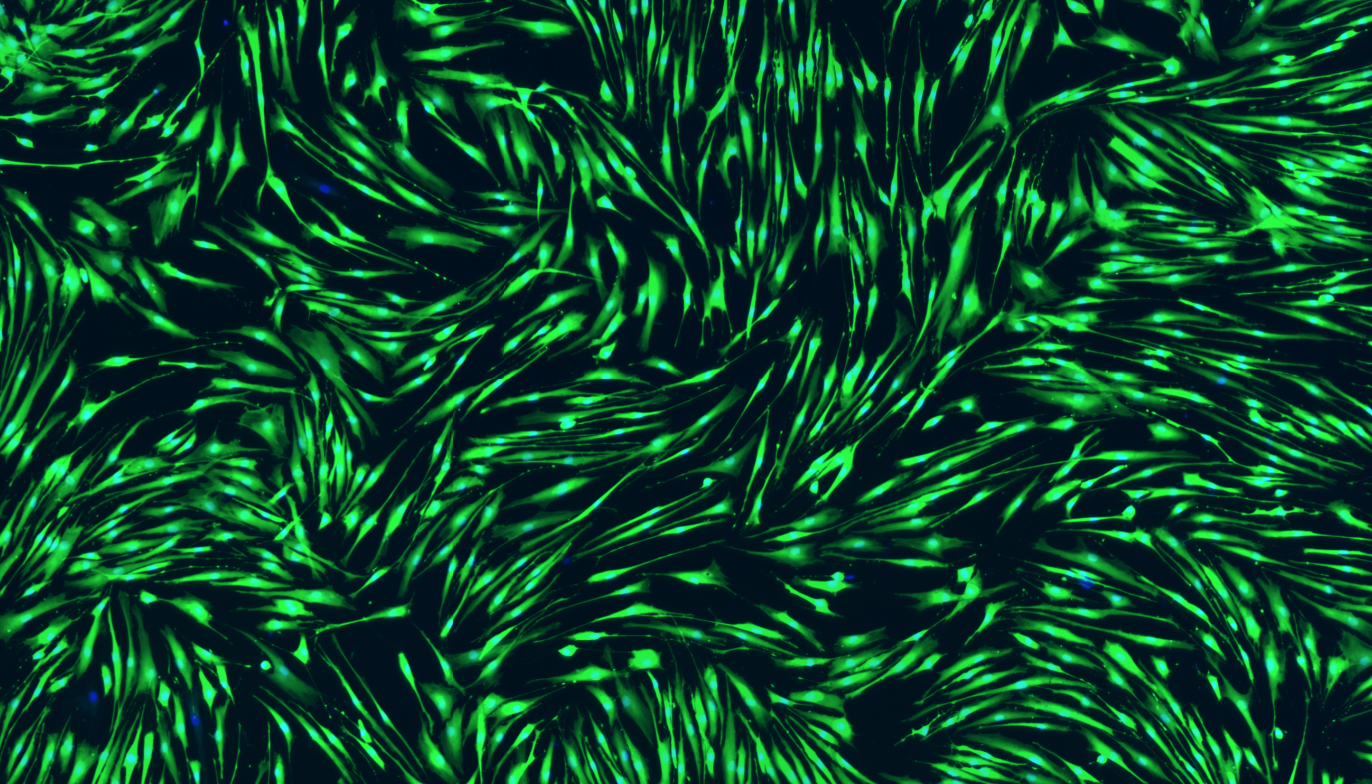
I’ve never worked with these cells before so, again, I needed to work out how many I’ll need to get a good result from the assay. These cells however grow as a very thin sheet which is so see-through that I initially couldn’t tell if the wells were full. To solve this I used stains called Calcein AM and Hoechst. Calcein AM can stain any living cell because they metabolise it from a see-through molecule into a fluorescent green product, whereas Hoechst can stick to DNA and turn all the cell nuclei blue, and so suddenly, all of my cells glow:
Aren’t they pretty?
Once they’re this easy to see, it’s even easier to see how many cells are needed to fill a well, but not get overcrowded. You can read more about how I did that in my Zenodo post.
The only question I have left is why some cells didn’t stain with Calcein AM, even though their nucleus looks fine? Anyone know the answer?

Hi Elizabeth! I am working on developing new inhibitors targeting Alk2. I however do need help choosing the correct cell lines. Do you think we chat sometime?