I’ve not posted in a while, mainly because things have been really busy, but good busy, as I now have 3 new datasets/structures, of ALK2 with M4K2118, M4K2149 and, amazingly, M4K2009. As you know, this is our lead candidate, which until now has not behaved very well in crystal trials, and it hasn’t been possible to get decent crystals. In my last post I told you how I had somehow managed to mix up the TNIK and ACVR1/ALK2 tubes when I was setting up the plates, and accidentally put ACVR1 into a coarse screen and TNIK into a fine screen for ACVR1. Well, after I got lovely big crystals for what I thought was ACVR1 with M4K3003, I sent them to the synchrotron, got beautiful data to 1.4 Å, phased it, and lo and behold! It was M4K2009 snugly fitted into the binding pocket (see the ligand density after molecular replacement below). Of course, I didn’t notice this was a trimethoxy containing compound (how??) before I excitedly emailed everyone to tell them I had M4K3003, and then had to retract … However, the story ends well as M4K2009 is really important.
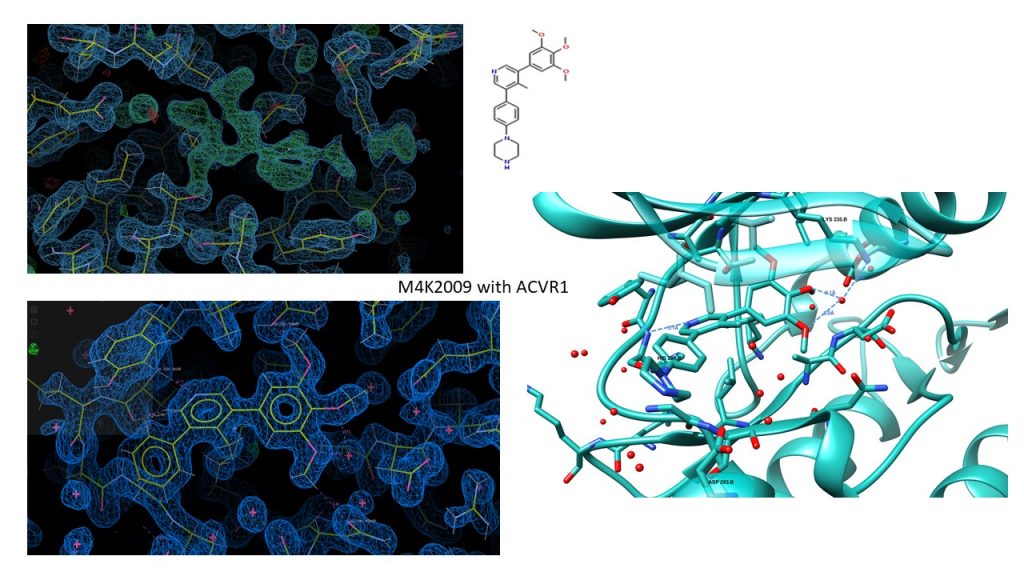
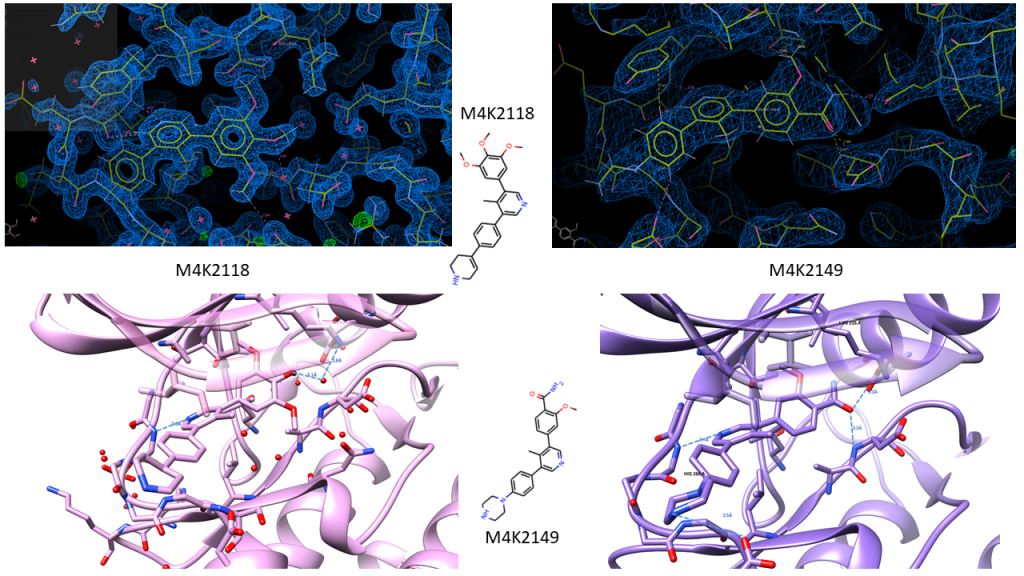
Additionally, I got data for M4K2149 (~2.6 Å), one of the amide head group containing compounds we’re interested to see, and M4K2118 (~1.4 Å), which is doing well in some of the other assays too. I’ve put the .mtz files here on Zenodo if you want to play with high resolution data, and some images below showing the ligand density and the ligands binding in the pocket. The amide head group of M4K2149 appears to preserve the water network in the pocket that we’ve observed in other structures, but as the resolution is too low for waters, we can’t tell til we get higher resolution data. Which hopefully will be this week, as I’ve grown some really pretty crystals for M4K2149 that I hope will diffract better. These appeared quite by accident again, as I’d designed a fine screen around the D5 well condition of my mega composite screen, but neglected to notice when I validated the screen that I’d put 10 x less ammonium sulphate in one row than intended – should have been 0.9 M, but ended up being 0.09 M. I should probably start calling myself ‘the accidental crystallographer’ at this rate.
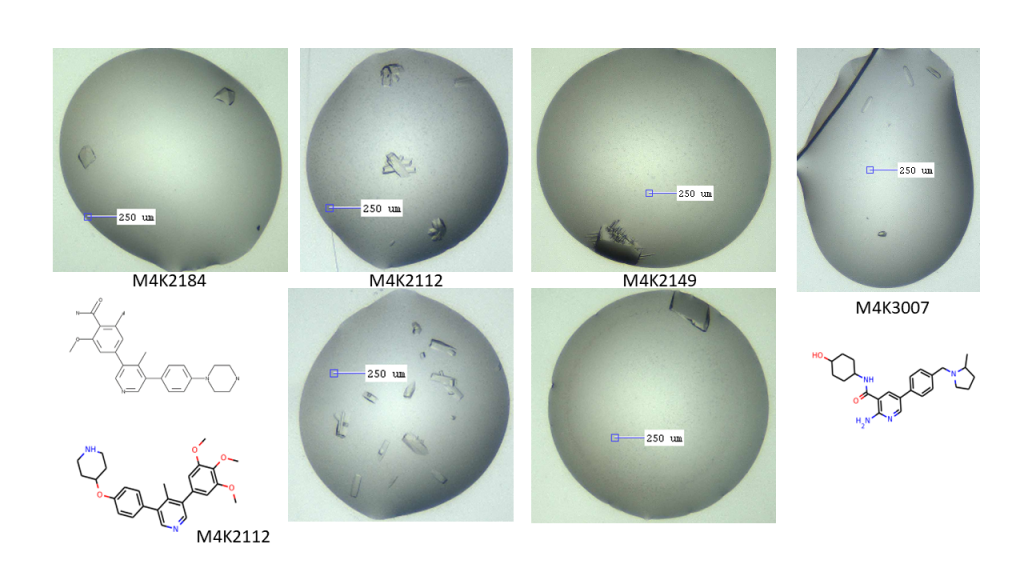
In the meantime, I’ve got some small crystals for M4K3007, another structure we really want, and some pretty ones for M4K2184, another amide head group compound. I’ve followed up the M4K3007, just waiting to see if anything appears, and will send the tiny ones to the synchrotron while waiting.
I’m also very happy to say that our new M4K2117 crystal system for XChem fragment screening has worked very nicely so far, in the sense that the crystals are large and relatively robust and can handle soaking in low salt conditions as well as compounds with DMSO for a few hours. They then still diffract to around 1.2-1.8 Å, consistently in the same space group, C 1 2 1. This is a vast improvement over the previous system with LDN-193189. We did a solvent characterisation and pre-screen a couple of weeks ago with 100 crystals, and of the 20 or so hits we got, none was in the allosteric pocket we are looking for. This could be because of the 1.4 M ammonium sulphate and 0.2 M Na/K tartrate I have in there, which might prevent access of fragments to the pocket. Thus, using the remarkable shifter that Nathan Wright at SGC designed, we’ll be transferring loads of crystals from grow plates to low salt soak plates before doing the compound soaks this week, with the same 100 compounds, in order to check if the hit rate/location changes.
I will mention though, for anyone who may never have noticed this, the DMSO concentration in the protein solution for crystallisation is very important! I had previously been using Batch 1 of M4K2117 for my co-crystallisation, but it ran out so I used Batch 2, which was a 50 mM stock, as opposed to the previous 25 mM stock. The plates I set up with this, the same in every other way, gave showers of rather ugly, crunchy, small crystals. So I tried a few things – diluted the stock to 25 mM in DMSO and set up plates with that, with both lower and higher concentrations of protein (8 and 14 mg/ml along with the usual 10-11 mg/ml), as well as using the stock as is, with lower and higher protein concentrations. The lower stock concentrations brought back the big, perfect, beautiful crystals from before, the higher stock, no matter the protein concentration, gave ugly chunks and showers. The final concentration of compound was the same in all, so clearly the DMSO percentage is a very important factor.
