ZFYVE9C (Otherwise known as SARA) is a scaffold protein involved in the association of various protein components as part of the TGFB signalling pathway. The equivalent in the BMP signalling pathway is a protein known as Endofin so studying SARA (which is slightly easier) might give us useful insights for both proteins. For more details on this project please see this blog post.
One of the unanswered questions about SARA is what does one of the major domains that it has actually do? (The domain is known as a DUF domain, or a Domain of Unknown Function!) We know the structure but that hasn’t given us any clues as to its function. One thought was that it might be involved in recruiting type I or type II receptors close to their binding partners such as one of the SMADs to allow signal propagation.
I purified the DUF domain of SARA and mixed it with either excess TGFBR1 or excess TGFBR2 (the two relevant receptors in the TGFB pathway). I tested the receptors in both the unphosphorylated (inactive) and phosphorylated (active) states. After incubating the samples for 30 minutes while concentrating the volume back down to <5ml, I ran them down a gel filtration column to see if the two proteins bound together sufficiently to shift the peaks coming off the column which would indicate they were acting as a complex of larger mass.
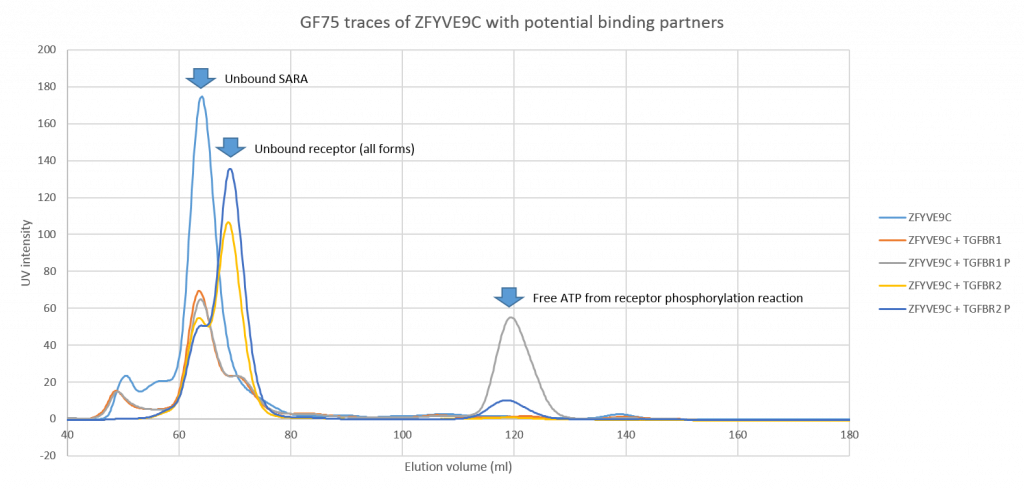
Overlay of the UV traces from all the size exclusion gel filtration runs to show the relative elution volumes of the peaks confirming no shift in the ZFYVE9C peak consistent with no complex being formed.
Unfortunately I didn’t see any shift in the peak for SARA suggesting that either the two proteins don’t bind or that the affinity is weak enough that they fall apart under the conditions of the gel filtration column.
The next thing to try will be to purify the proteins again but not to cleave the tag on one of the binding partners (either the receptors or the SARA protein) and try to do a pull down experiment to test whether this shows any association or not.
You can find more details over at Zenodo.

If it were me, I’d drop it and think of a more agnostic (unbiased) way to test its function
Your approach is almost always a losing game, since a negative result is meaningless and it takes years to gain confidence in any interaction (since you “forced” the proteins to interact, you’d need to go back into cells, do mutants etc)
so why not just start with the cell experiment and ask “what co-purifies with it”?
That’s a fair point – I think my hope with at least the initial experiment was that if I had seen an association (and given it’s presumed function in the pathway it wasn’t an impossibility and I’d have felt very foolish not to at least try such an easy experiment) I could have instantly set it up in crystal plates which would have been very interesting.
There have been a few studies indicating a few other potential binding partners including TOM1 which I’ve looked at in the past and while I’ve been able to detect an interaction by ITC I’ve not been able to get a crystal structure. The fold of the DUF domain is similar to the fold of SUFU in the hedgehog pathway but there is very little sequence similarity so it’s hard to predict how the binding site and substrates of SUFU will map to ZFYVE9C.
In cell studies is something to consider too. It’s not where my expertise lies but I’m hoping to talk to Jong Fu about whether he’s got any thoughts on it.