You may recall I was in Montreal for the last three months doing an internship with Molecular Forecaster Inc. (MFI), a computational chemistry company. I’m back in Toronto from my internship, ready to jump back into my research, but first I’ll summarize the virtual screening work I did at MFI for USP5.
In a previous post, I assessed MFI’s FITTED docking platform for ranking success on a library of experimentally tested compounds and found that flexible docking with MATCHUP resulted in the best overall ranking of USP5 ZnF-UBD compounds. I used MFI’s flexible protein docking with MATCHUP to screen large commercial libraries. You can see details on Zenodo.
First, I selected commercial libraries of compounds from reliable vendors from the ZINC15 database, for at total of 607, 580 compounds. I then used REDUCE to filter the chemical library to 505, 772 compounds and SELECT to create a subset of the library by removing previously screened or experimentally tested compounds for a total of 225, 665 compounds. Finally, I used FITTED with MATCHUP to dock the ligands to USP5 ZnF-UBD. Since the library of compounds I was screening was large (>225K compounds), I relied on the docking scores generated by FITTED to compile a list of 347 ligands that had the best docking scores (Figure 1).
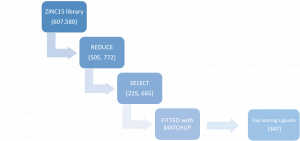
Figure 1. Summary of virtual screening workflow
A large majority of the top scoring ligands are small fragments, with carboxylate groups, which is not surprising considering the carboxylate group mimics the native C-terminus of ubiquitin which binds to the USP5 ZnF-UBD. I will look through the list of top scoring ligands and determine if there are any favorable scaffolds with new binding pocket interactions that might improve potency. For example, ZINC000000376528 makes hydrogen bonds with Y261, R221 backbone and side chain, similar to hits we have seen before (PDB: 6DXT, 6NFT, 6DXH). Interestingly, an amino group makes hydrogen bond interactions with D264. It is possible that this additional hydrogen bond interaction, which has not been seen in previous hits could improve potency. It may also be possible to engineer existing hits with this group, if this is the case.
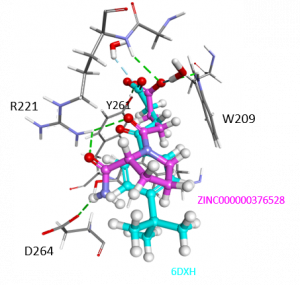
Figure 2. Docked pose of ZINC000000376528 (magenta) and co-crystallized ligand PDB: 6DXH (cyan)
This week will be busy, planning and organising future work as well as selecting potential new scaffolds for ordering. I also have the University of Toronto’s 2019 Visions in Pharmacology (VIP) research day this week, for which I’ll be doing a poster presentation on my work with USP5. I’m looking forward to hearing about the research being done by other students in my department. Stay tuned!
| Structure Image | Smiles | Mol. Weight |
|---|---|---|
| O=C(O)[C@@H](O)[C@H](O)[C@@H](O)[C@@H](O)CO | 196.2 | |
| O=C([O-])[C@H](O)[C@H](O)CO | 135.1 | |
| OC[C@@H]1O[C@@H](O)[C@H](O)[C@@H](O)[C@@H]1O[C@@H]1O[C@H](CO)[C@@H](O)[C@H](O)[C@@H]1O | 342.3 | |
| N[C@@H](C[C@@H](O)C(=O)O)C(=O)O | 163.1 | |
| N[C@H](CCS(=O)(=O)O)C(=O)O | 183.2 |
