I joined the group of Dr. Cheryl Arrowsmith 2 months ago. Currently, I am working on the Huntington’s disease project along with Dr. Rachel Harding and Jacob McAuley. During these past two months, I have been learning how to purify huntingtin (HTT) protein and overall getting up to date with the project.
A brief introduction to the project: HTT is a 348 KDa protein that is vital for development 1. HTT has a polyglutamine repeat at its N-terminus which is normally no longer than 35 repeats (Figure 1). When this number of repeats exceeds 38 repeats, it causes Huntington’s disease (HD), a dominantly inherited neurological disorder that occurs in adulthood and sometimes in children 2. Thus, like many other neurological disorders such as Parkinson’s and Alzheimer’s disease, HD is also caused by a protein with an abnormal function in the cell. There are very limited treatments available for HD patients in part because there is little information about the function and structure of HTT and mutant HTT.
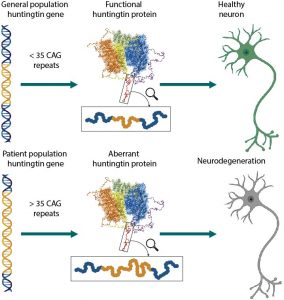
Figure 1. The huntingtin protein in normal population and in Huntington’s disease patients. Figure courtesy of Dr. Rachel Harding.
In the cell, HTT is thought to be involved in several important protein-protein interactions that are critical for cellular vesicular transport, endocytosis, autophagy and the regulation of transcription 3,7. However, the complete picture of the function of HTT in the cell is yet not fully determined.
HTT is made up of three different domains. Two HEAT 8,9 (Huntingtin, Elongation factor 3, protein phosphatase 2A, Target of rapamycin 1) domains at the N-terminus and C-terminus which are connected by a bridge. However, HTT has several disordered regions within these domains, which makes structural studies of HTT challenging. Recently, the cryo-EM structure of the protein was obtained making a huge step forward to understand more about HTT 10. However, almost a quarter of the structure of HTT is still unknown in part due to the dynamic nature of this protein.
The main focus of my project is to obtain the 3D structure of the individual domains of the HTT protein, namely the N-terminal HEAT domain, the bridge and C-terminal HEAT domain. With this approach, we hope to better define structural elements in the protein. Further, studies with the individual domains will also be useful to understand what the role of each of the domains is within HTT. By learning about the structure and function of the individual domains, we can learn about HTT’s function inside the cell. I will be posting updates on the research progress so stay tuned!
1. F. Saudou and S. Humbert. Neuron, vol. 89, no. 5, pp. 910-926, doi: https://doi.org/10.1016/j.neuron.2016.02.003.
2. M. E. MacDonald et al., ” Cell, vol. 72, no. 6, pp. 971-983, 1993/03/26/ 1993, doi: https://doi.org/10.1016/0092-8674(93)90585-E.
3. F. Mochel and R. G. Haller. (2011)The Journal of Clinical Investigation, vol. 121, no. 2, pp. 493-499, doi: https://doi.org/10.1172/JCI45691.
4. P. Harjes and E. E. Wanker. Trends in Biochemical Sciences, vol. 28, no. 8, pp. 425-433, 2003/08/01/ 2003, doi: https://doi.org/10.1016/S0968-0004(03)00168-3.
5. S.-H. Li and X.-J. Li, The Neuroscientist, vol. 10, no. 5, pp. 467-475, 2004/10/01 2004, doi: https://doi.org/10.1177/1073858404266777.
6. J. P. Caviston and E. L. F. Holzbaur. Trends in Cell Biology, vol. 19, no. 4, pp. 147-155, 2009/04/01/ 2009, doi: https://doi.org/10.1016/j.tcb.2009.01.005.
7. Z. Chiara and C. Elena. Oxford, UK: Oxford University Press.
8. M. A. Andrade and P. Bork, Nature Genetics, vol. 11, no. 2, pp. 115-116, 1995/10/01 1995, doi: 10.1038/ng1095-115.
9. J. Perry and N. Kleckner, Cell, vol. 112, no. 2, pp. 151-155, 2003, doi: 10.1016/S0092-8674(03)00033-3.
10. Q. Guo et al., Nature, vol. 555, no. 7694, pp. 117-+, Mar 2018, doi: 10.1038/nature25502.
