There are two major NSD3 isoforms expressed, long (aa 1-1437) and short (aa 1-645, differing in sequence from 620-645). The short isoform lacks a methyltransferase domain and, perhaps surprisingly, is the form attributed to NSD3’s role in acute myeloid leukemia (AML) [1]. I’ve previously used the TCGA-LUSC data-set [2] to test for differential expression based on the abundance of NSD3 transcripts at the gene-level. Next, I was interested in using the isoform-level expression data from Firebrowse to look at how different isoforms may be regulated. Is expression correlated or are there more complex regulatory mechanisms that control splicing decisions?
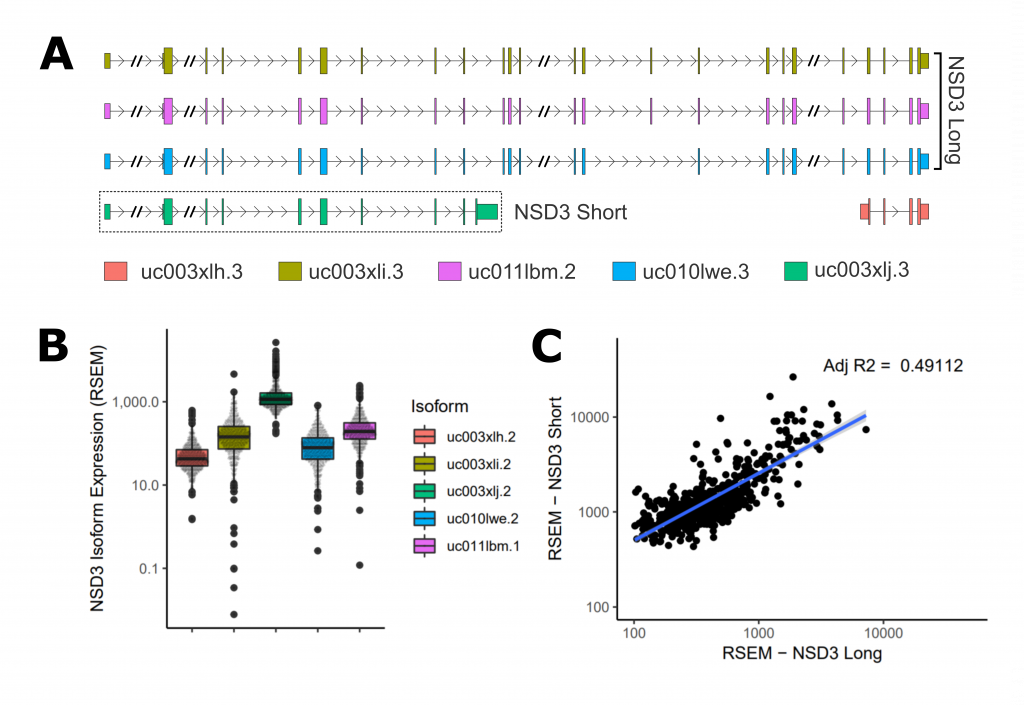
Figure 1. Isoform Expression Analysis (A) Schematic representation of NSD3 isoforms. Introns lengths have been shortened in several places for visualization. (B) Normalized expression levels of NSD3 isoforms (RSEM). (C) Relative expression of short vs long isoforms.
Firstly, I found that the short isoform is expressed roughly 10-fold over long isoforms, making it the dominant isoform in the LUSC data (Figure 1 – B). Next, when comparing transcript levels of long vs short, I observed a significant moderately positive correlation, suggesting that the isoforms are likely co-regulated. I hope the analysis will be helpful in framing the shared and separate functions of the two isoforms uncovered in future experiments.
As always, the complete analysis & code can be found on zenodo.
Acknowledgement: The results shown here are in whole or part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
References
1. Shen C, Ipsaro JJ, Shi J, et al. NSD3-short is an adaptor protein that couples BRD4 to the CHD8 chromatin remodeler. Molecular cell. 2015;60(6):847-859. doi:10.1016/j.molcel.2015.10.033.
2. Weinstein JN, Collisson EA, Mills GB, et al. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nature genetics. 2013;45(10):1113-1120. doi:10.1038/ng.2764.
