Chemical probes typically bind off targets in addition to their intended target protein. Since it is hard to determine whether the phenotype is due to inhibition of the target protein or off-targets, the use of negative controls, which are structurally close to the probe but are inactive against the intended target, is highly recommended. Loss of a chemical probe associated phenotype upon treatment with negative control increases the confidence that the phenotype is elicited by the activity of the chemical probe at the target protein.
When there is a chiral center in a chemical probe, a common practice to generate the negative control is to purify and test both enantiomers. If the mirror enantiomer of the chemical probe is inactive on the intended target, it is designed as the negative control.
However, if enantiomer A (the probe) is active against unknown off-targets the risk that enantiomer B (the negative control) is inactive against these off-targets is unknown (Figure 1).

Figure 1: enantiomer B can be inactive against a protein X if enantiomer A is active against the same protein X.
My research goal is to systematically evaluate how often enantiomer B as negative control is inactive against a protein X if enantiomer A is active against the same protein X (Figure 2).
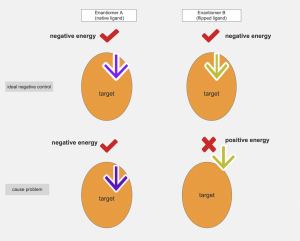
Figure 2: when both enantiomer A (native ligand) and enantiomer B (flipped ligand) can bind off target proteins with negative energy, the flipped ligand can be used as an ideal negative control. When enantiomer B (flipped ligand) cannot bind off target binding pocket since there are steric clashes, the flipped ligand should not be use as negative control.
Given a crystal structure of one enantiomer bound to a protein, we can calculate the binding energy of the native ligand (enantiomer A) for protein X and the energy of the “flipped” ligand (enantiomer B) for the same protein X. A positive energy of the flipped ligand indicates that there are steric clashes and that the ligand is inactive against this protein. Applying this approach to the hundreds of structures of chiral ligands in the Protein Databank (PDB) can indicate the likelihood that the mirror enantiomer of chemical probe does not bind to off targets of the chemical probe. If you would like to know more detail descriptions of methods and script, please refer to my Zendo report.
After flipping the chiral center of these compounds, we can calculate the energy between each ligand and its binding pocket. A positive flipped energy indicates that there are strong steric clashes of the flipped ligand with the binding pocket which abrogates binding to the target. There are 20% compounds with negative native energy and positive flipped energy, indicating that while these ligands are active against a target protein, its enantiomer are inactive against the same target.
Together, these data indicate that about 80% of off-targets inhibited by a chemical probe containing a chiral center will also be inhibited by a negative control consisting of the mirror enantiomer of the chemical probe. The common practice of using as a negative control a mirror enantiomer when available seems therefore justified, though not risk-free.
Based on computational analysis of over 1000 chiral drug-like ligands bound to proteins in the PDB, we find that mirror enantiomers will retain binding to the same target protein in 80% of cases. The difference in binding potency between enantiomers should be evaluated to complete this analysis.
