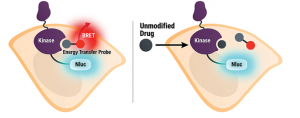
In my previous post, I presented different CLK2 inhibitors (https://openlabnotebooks.org/clk-chemical-probe/), including structures, and IC50 curves that were generated using a cell target engagement assay called NanoBRET. My colleagues Carrow Wells and Julie Pickett generated these data. The BRET of NanoBRET stands for bioluminescence resonance energy transfer, and it is an in-vitro quantitative technique to identify target engagement of the drug with the protein of interest. As shown in the picture below, the kinase has to be modified somehow to express the NanoLuc, used as a detector. Next, a promiscuous and fluorescent tracer has to be identified and be introduced to engage with the kinase, which results in a BRET signal. Finally, our test compound, hopefully a selective inhibitor (called the unmodified drug in the picture) is introduced, resulting in a decrease of the BRET signal, due to displacement of the fluorescent tracer from the active site of the tagged kinase.
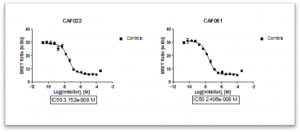
Compounds CAF022 and CAF061, previously identified as CLK2 inhibitor candidates, were tested using NanoBRET cell engagement. IC50 curves were generated with 10 different inhibitor concentrations, providing an IC50(CAF022) = 32 nM and IC50(CAF061) = 24 nM, demonstrating high affinity CLK2 target engagement in cells.
In order to meet the SGC criteria and compounds become a chemical probe, potency is not enough and other chemical/biological factors have to be addressed. Selectivity is a key factor, and the requirement is a >30-fold difference within the family. We are currently assessing kinome wide selectivity of our top candidates. This involves a kinome scan at Discoverx (>400 kinases) testing our inhibitors at a concentration of 1 micromolar.
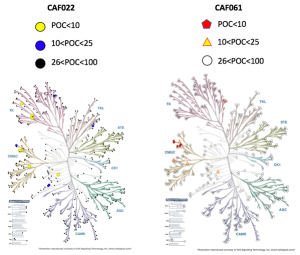
The picture below shows the kinome scan results of both CAF022 and CAF061.
Disclosure of CAF022, this compound is already pretty selective, hitting 8 kinases with >90% of remaining activity, containing 3 false positives (checked by Kd’s follow-up). These 5 kinases include: PIP4K2C, MAPK15, CLK1, STK16 and CLK2 (yellow dots). Furthermore, between 89-75% of remaining activity, only 6 kinases (2 of them false positive) were hit, including NEK6, NEK7, HIPK1 and TGFBR2 (blue dots).
Regarding CAF061, this compound shows a cleaner kinome scan, hitting only 5 kinases with >90% of remaining activity (Kd value determination is in progress to check on false positives). These kinases included: HIPK1, HIPK2, CLK1, CLK2 and CLK4 (red pentagons). An extra 5 kinases were found to have a remaining activity between 89-75%, including: HIPK3, STK16, CDKL3, ERK8 and JAK1 (JH2 pseudo-kinase) (yellow triangles). Once again Kd determination to determine true off-targets is in progress.
Having found common kinases off-targets, Kd values will be generated for those and other potential off targets and/or in a similar nanoBRET cellular assay to get IC50. Having found a selective compound, then we are a step closer to a potential chemical probe. Feel free to suggest further ideas and if you have any questions or comments or want more detail on anything, please let me know and feel free to contact me or any of my SGC colleagues.

Carla, could you explain the readout of Kinome Scan? Is it the % of inhibition relative to a potent inhibitor serving as positive control?
Matthieu, DiscoverX use immobilized promiscuous kinase inhibitors. Our test compounds compete for the active site and displace ligand if they are good inhibitors. If our compounds are no good they act like DMSO/buffer and show up as 100% of control. More info: https://www.discoverx.com/technologies-platforms/competitive-binding-technology/kinomescan-technology-platform
Carla, it looks like you are close to having a chemical probe for CLK2. How do the best compounds compare with other known CLK2 inhibitors?
Tim, there are inhibitors out there. For example TG003 which has an IC50 of 20 nM, 200 nM, and 15 nM for CLK1, CLK2, and CLK4, or T3, a compound from Takeda Pharmaceutical, with an IC50 of 0.67 nM and 15 nM for CLK1 and CLK2 (find more info here: DOI 10.1038/s41467-016-0008-7). The next step is going to be the generation of the IC50 curves for CLK1 and CLK4, to see if we can actually make a difference between the family.