In my last post, I analyzed a structure of USP5 zinc-finger ubiquitin binding domain with a cysteine-glutathione disulphide bond. These crystals were not suitable for ligand soaking due to the formation of a disulphide bond at Cys195 with a glutathione additive that occupies the ubiquitin binding pocket in the adjacent molecule in the crystal lattice.
I went back to the base crystallization condition and optimized the pH and concentration of components of the crystallization buffer. Experimental details can be found on Zenodo. I determined a condition of: 1.5 M ammonium sulfate, 0.1 M sodium cacodylate pH 6.8, 0.2 M sodium acetate, 2% glycerol that resulted in consistent and uniform three-dimensional crystals that were reproducible and diffracted well.
The structure and how it was solved can be found here. The structure of the USP5 Zf-UBD has the same space group and unit cell dimensions as the already solved apo structure (PDB: 2G43) and the zinc-finger ubiquitin binding domain is next to a solvent channel suggesting the crystal form is amenable to soaking with small ligands. Additionally, my crystals diffract to higher resolution (1.6 Å vs. 2.1 Å), allowing me to resolve more structural features of the protein and future protein-ligand complexes.
X-ray crystallography is done at cryogenic temperatures to limit radiation damage and requires cryo-protection to cool the crystals to cryogenic temperature without ice formation and damage to the crystal1. The small ligands that will be used for soaking with this crystal form are prepared as 200 mM stock solution in DMSO. The presence of a solvent such as DMSO can help to increase ligand solubility as it is not volatile and is able to dissolve both polar and non-polar compounds2. For these reasons, it was important to optimize the cryo-protectant conditions and assess the tolerance of the crystals to DMSO for the USP5 Zf-UBD crystals. The crystal form was partially cryo-protected with 10% glycerol and was tolerant of 1% DMSO conditions (Figure 1). Soaking with ligands in 1-5% DMSO should be sufficient for compounds screening.
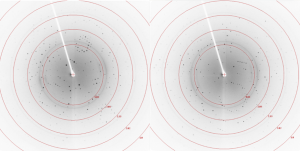
Figure 1. Diffraction images with USP5 Zf-UBD crystals in 1.5 M ammonium sulfate, 0.1 M sodium cacodylate pH 6.8, 0.2 M sodium acetate, 2% glycerol soaked in 10% glycerol, 1% DMSO for 30 minutes
Next, I’ll have to increase the concentration of the cryo-protectant to avoid the formation of ice rings (as seen in Figure 1), which can cause damage to the crystal. I hope to begin soaking experiments with small ligands once I’ve finished the optimization of the cryo-condition.
1 Farley, C. & Juers, H. D. Efficient Cryoprotection of Macromolecular Crystals using Vapor Diffusion of Volatile Alcohols. J Struct Biol, 2014. 188(2): 102-106.
2 Muller, I. Guidelines for the successful generation of protein-ligand complex crystals. Acta Crystallogr D Struct Biol, 2017. 73(2): 79-92.
