In two of my previous posts 9 and 10, I showed how we found the residues lining the catalytic pocket of SARS-CoV-2 Papain-like protease (PLPro) and we looked at the variability of this site across Alpha- and Betacoronavirus entries from UniProt.
In addition to the UniPro sequences, we were interested in looking at variants from COVID-19 patients. Nicola De Maio, our collaborator from Nick Goldman’s lab at EBI, looked at more than 15000 SARS-CoV-2 samples and identified all mutations at the PLPro catalytic site. Nicola saw five non-synonymous variants in his sample batch as shown in table 1.
For example, he saw that at residue number 247 of PLPro where the wild-type residue is proline, there were 3 non-synonymous mutations: 1) proline to serine (P247S) 2) proline to leucine (P247L) 3) proline to glutamine (P247Q).
| Residue | Non-synonymous variations at PLPro catalytic site |
| P247 | P (15868) S (1), L (2), Q (1) |
| P248 | P (15871), S (1) |
| T301 | T (15864), A (5) |
Table 1. The non-synonymous variants at PLPro catalytic site across more than 15000 SARS-CoV-2 samples.
What is important about these variants which come from COVID-19 patients is their possible effects on ligand-binding. To predict the effect, we modeled those five mutations from table 1 in ICM (Molsoft, SD) and used the change in Gibbs free energy (ddGbind) to predict the effect of those mutations on ligand binding.The ligand, in this case, is the viral inhibitor, VIR251 (PDB: 6wx4).
In Table 2, the ddGbind values for all five non-synonymous mutations are shown.
| Residue Number | Wild Type | Mutant | ddGbind | dGbind Wild type | dGbind Mutant |
| 247 | pro | leu | 5.2 | -34.2 | -29.0 |
| 247 | pro | ser | -0.5 | -34.2 | -34.7 |
| 247 | pro | gln | 0.7 | -34.2 | -33.5 |
| 248 | pro | ser | 3.5 | -34.2 | -30.7 |
| 301 | thr | ala | 0.0 | -34.2 | -34.2 |
Table 2. Energy calculations of non-synonymous mutations at the SARS-CoV-2 PLPro catalytic site to predict their effect on the binding of VIR251. The energy unit for ddGbind values is Kcal/mol.
Looking at table 2, we see that ddGbind values for P247L, P248S are above 2Kcal/mol which is considered a significant change in binding according to a previous study.1 P247S, P247Q, and T301A were predicted to have minor effects on VIR251 binding (-2kcal/mol < ddGbind < 2kcal/mol).
Using ribbon diagrams in Figures 1 and 2, you can see how the modeling in ICM works.
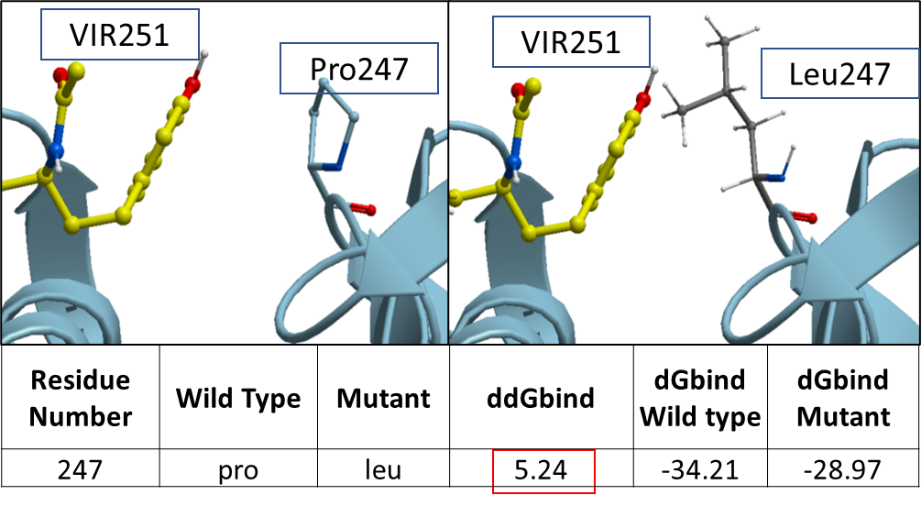
Figure 1. The ribbon diagram of SARS-CoV-2 PLPro’s variant at P247L.
We think that since leucine is a bulkier residue compared to proline, the mutation from proline to leucine has a destabilizing effect on the inhibitor binding.
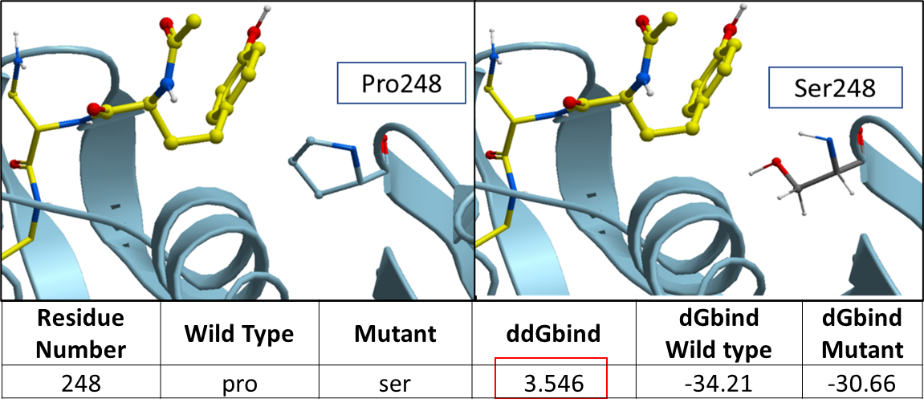
Figure 2. The ribbon diagram of SARS-CoV-2 PLPro’s variants at P248S.
The ddGbind value corresponding to P248S is predicted to be at 3.547 Kcal/mol. Proline is a non-polar aliphatic residue which makes hydrophobic interaction with the aromatic ring of Tyrosine on VIR251. The change to serine which is a polar non-charged residue may negatively impact hydrophobic interactions with VIR251.
In figure 10 of my Zenodo report, you can find the 2D ligand-protein diagram which highlights the chemical interactions between VIR251 and SARS-CoV-2 PLPro.
Lastly, the color-coded structure in figure 3 highlights the variants among the SARS-CoV-2 samples at PLPro catalytic site.
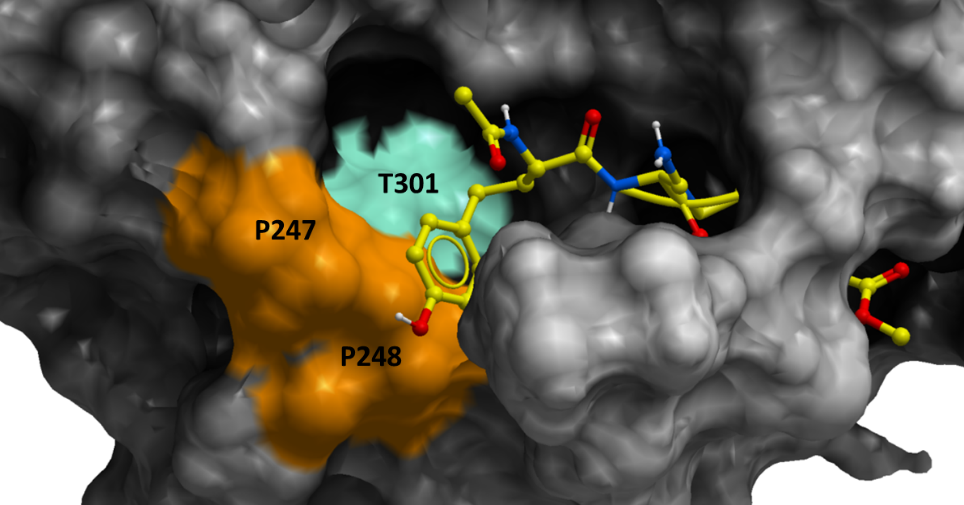
Figure 3. Color-coded surface representation of SARS-CoV-2 PLPro. The colored areas on the mesh show the non-conserved residues across more than 15000 SARS-CoV-2 samples from COVID-19 patients. The orange-colored residues correspond to variants, P247L and P248S, which penalizes VIR251-binding significantly, while the green-colored residues are the mutations that does not penalize ligand binding significantly.
Altogether, the SARS-CoV-2 variants that we looked at are predicted to penalize VIR251-binding only slightly with SARS-COV-2 PLPro. That being said, our method has only a predictive power and the findings should be confirmed further in the lab.
In my next post, I will show you how we predict the effect of all possible mutations at the twenty-four sidechains of the PLPro catalytic site on VIR251-binding.
Please contact me via the “Leave a comment” link at the top of this post. Stay Tuned for more updates on this project!
Reference:
- Schapira, M., Totrov, M., Abagyan, R. Prediction of the Binding Energy for Small Molecules, Peptides and Proteins. Journal of Molecular Recognition 12(3), 177-90 (1999). https://doi.org/10.1002/(SICI)1099-1352(199905/06)12:3<177::AID-JMR451>3.0.CO;2-Z
