In my last post, I showed how we found the residues lining the catalytic site of SARS-CoV-2 Papain-like protease (PLPro).
In this post, I will show the sequence diversity across UniProt entries from the Alpha- and Betacoronavirus genera and map that to the PLPro’s catalytic site using its crystal structure (PDB: 6wx4).
In the context of the emergence of future MERS-like or SARS-like coronaviruses from the bat strains circulating in bat reservoir species, it is imperative to do a broad survey of viral proteins to identify the best strategies for the development of broad-spectrum viral inhibitors.1 For this exact reason, we are looking at twenty-eight reviewed sequences from Uniprot, where six belong to entries of Alphacoronavirus genus and twenty-one belong to the Betacoronavirus genus. We then assess their amino acid variability at PLPro catalytic site where the non-natural amino acid-containing inhibitor, VIR251 binds (PDB: 6wx4).
I made a diversity dendrogram focusing on the twenty-four amino acid residues of the PLPro catalytic site shown below in figure 1. Strikingly, across these twenty-eight entries, only five out of twenty-four residues at PLPro catalytic site are conserved: C111 (the catalytic residue which binds covalently to the inhibitor VIR251), D164, G271, H272, Y273. In other words, this catalytic site is highly variable among the entries we have looked at.
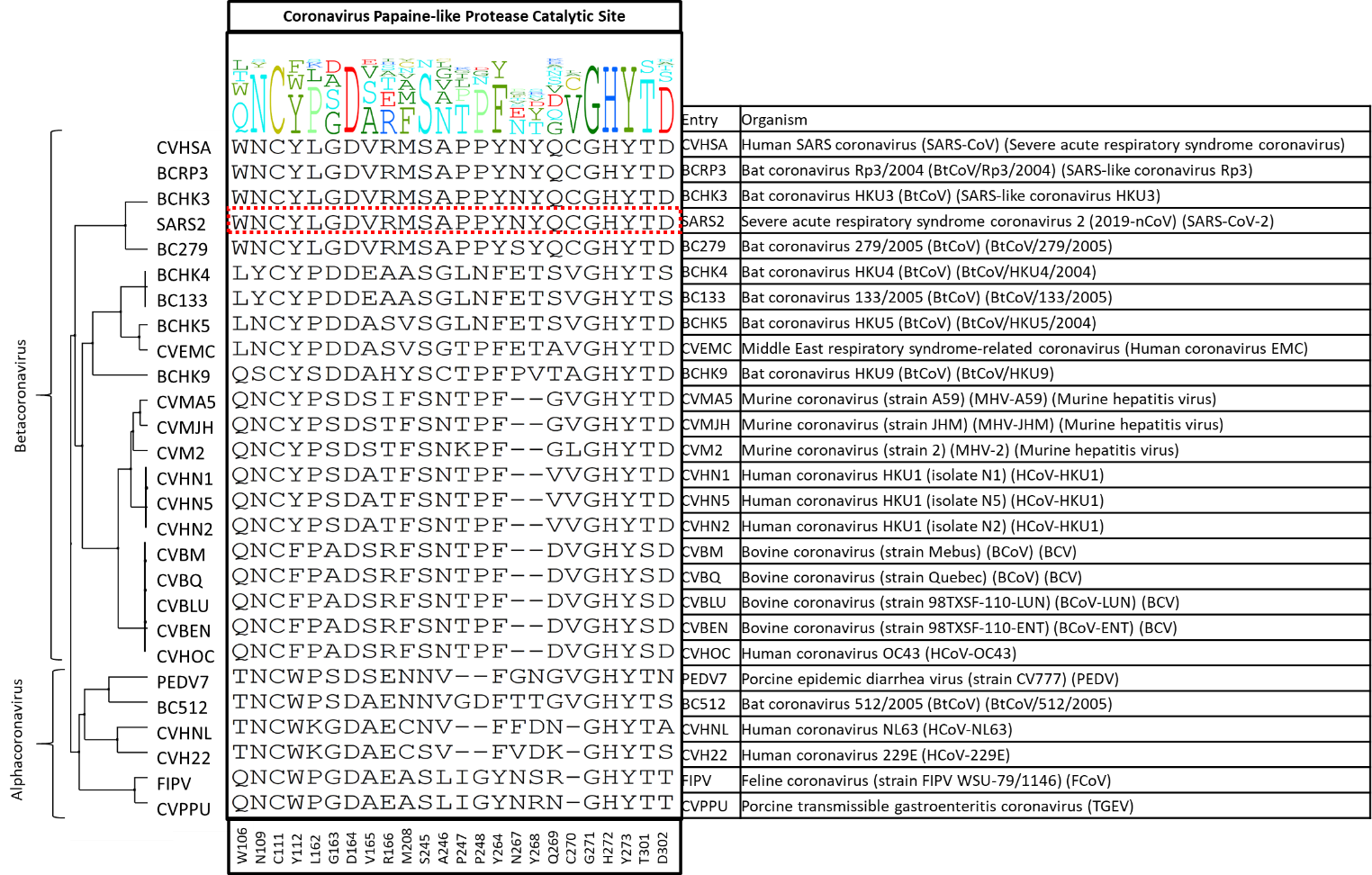
Figure 1. The diversity dendrogram of the catalytic site of PLPro for the entries of Alpha- and Betacoronavirus genera.
We then made a diversity dendrogram of the twenty-one entries of the Betacoronavirus genus. As shown in figure 2, we still observe high variability among these entries where only six out of twenty-four residues are conserved at PLPro catalytic site: C111 (the catalytic residue which binds covalently to the inhibitor VIR251), D164, S245, G271, H272, Y273.
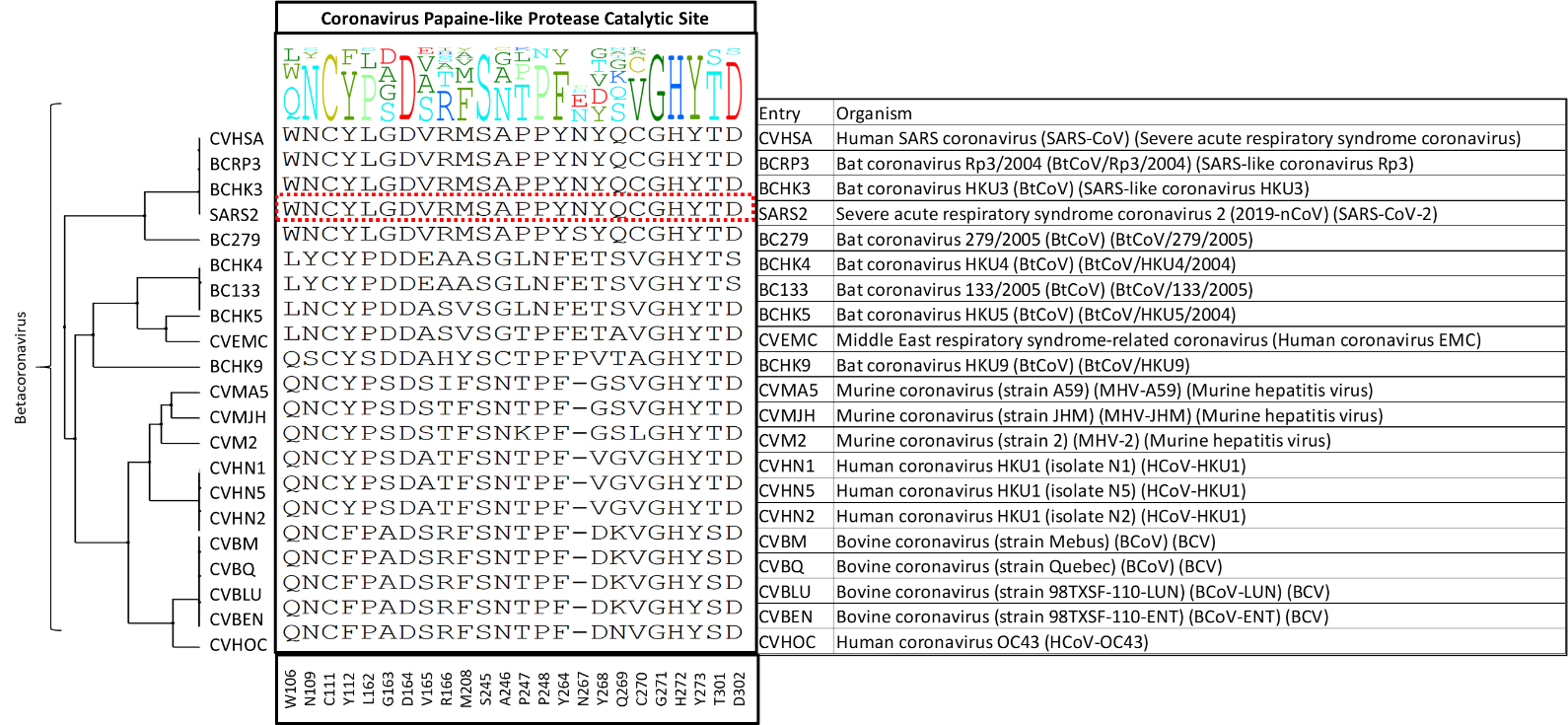
Figure 2. The diversity dendrogram of PLPro catalytic site residues among the UniProt entries of the Betacoronavirus genus.
To help you visualize the residues that varied among the twenty-eight entries of Alpha- and Betacoronavirus genera, I have made a color-coded surface representation of SARS-CoV-2 PLPro as shown in figure 3.
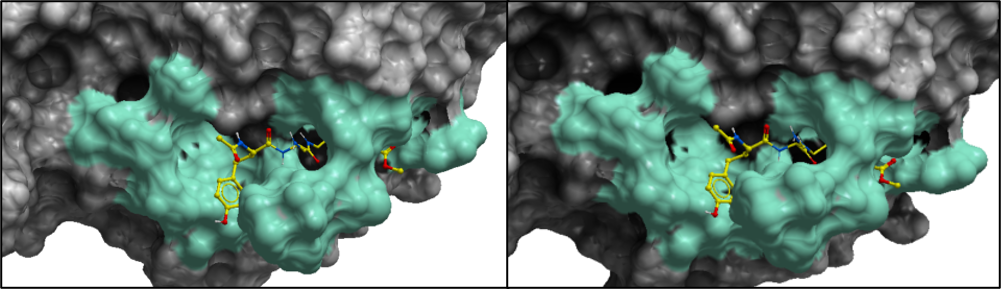
Figure 3. Mapping the genetic variation of entries in the Alpha- and Betacoronivirus genera onto SARS-CoV-2 Crystal structure (PDB:6wx4). The green color shows the non-conserved residues among the entries from the Alpha- and Betacoronavirus genera. On the left, all twenty residues that were non-conserved among the twenty-eight entries of Alpha- and Betacoronavirus genera are shown (W106, N109, N109, Y112, L162, G163, V165, R166, M208, S245, A246, P247, P248, Y264, N267, Y268, Q269, C270, T301, D302). On the right, all nineteen residues that were non-conserved among the twenty-one entries of Betacoronavirus genus are shown (W106, N109, N109, Y112, L162, G163, V165, R166, M208, A246, P247, P248, Y264, N267, Y268, Q269, C270, T301, D302).
In one of my future posts, I will cover the predicted effect of each variation mentioned above in terms of how it alters the binding of the inhibitor molecule, VIR251, to PLPro.
From the diversity dendrograms shown above, it became evident that the catalytic site residues of PLPro are mainly non-conserved and that would limit its application as a target-site for wide-spectrum anti-coronavirus inhibitors.
In my next post, I will show the energy calculations corresponding to the variants from SARS-CoV-2 patient samples at the catalytic site of PLPro.
To view the corresponding Zenodo report, please click here.
Stay in touch and leave your comments using the “Leave a comment” link at the top of this post. Stay Tuned for more updates on this project!
References:
- Sheahan, T. P. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Science Translational Medicine 12 (541). 2020. https://doi.org/10.1126/scitranslmed.abb5883
