Following our work on SARS-CoV-2 NSP16, we shifted our focus on SARS-CoV-2 Papain-like protease (PLPro) which is a cysteine protease.
A protease is an enzyme that breaks down proteins into smaller polypeptides or single amino acids. Similarly, PLPro recognizes specific tetrapeptide motif (LXGG) found in-between the fused viral proteins and cuts them at those particular sites similar to a scissor. Through PLPro’s function, nsp1, nsp2, and nsp3 proteins get released, which are necessary for viral replication.1
At the same time, PLPro also helps the virus evade the host innate immune responses through its effects on the deubiquitination and deISGylation process. Hence PLPro is essential for SARS-CoV-2 replication and is a promising target for antiviral drugs development.1
Wioletta Rut and their colleagues have crystalized structures of two inhibitors (VIR250 and VIR251) in complex with SARS-CoV-2 PLpro, which uncovers their inhibitory mechanisms (PDB: 6wx4).1 The Catalytic cysteine 111 of CoV-2-PLpro participates in a chemical reaction leading to the formation of a covalent thioether linkage between VIR251 and Cys111 of PLPro, as shown in figure 1.
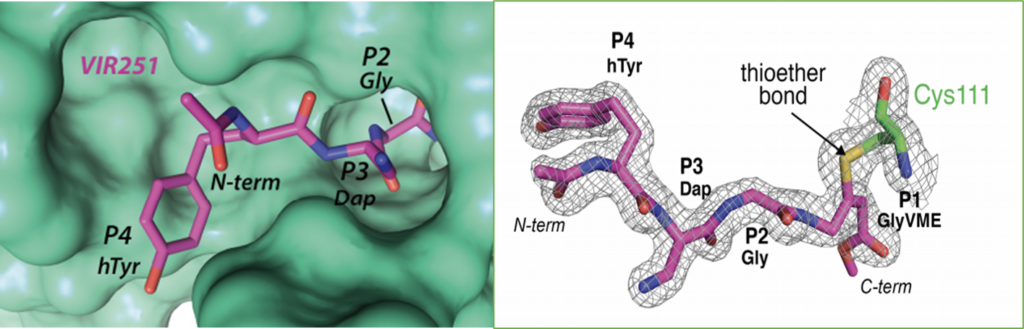
Figure 1. Formation of thioether bond between the non-natural amino acid-containing inhibitor, VIR251, and Cys111.1
In this post, I will explain how we identified the active site of PLPro and its druggability measure, and list the residues lining the catalytic side of PLPro.
Using ICM function PocketFinder (Molsoft, SD) we found the potential drug-binding sites using the PLPro x-ray crystal structure (PDB: 6wx4). Then using SiteMap (Schrodinger, NY) we assessed the druggability of the binding sites using the druggability score (Dscore). The catalytic site of PLPro has a Dscore of 0.81 indicative of a difficult druggable site. At the same time, this site has shown to be druggable using covalent inhibitors.1
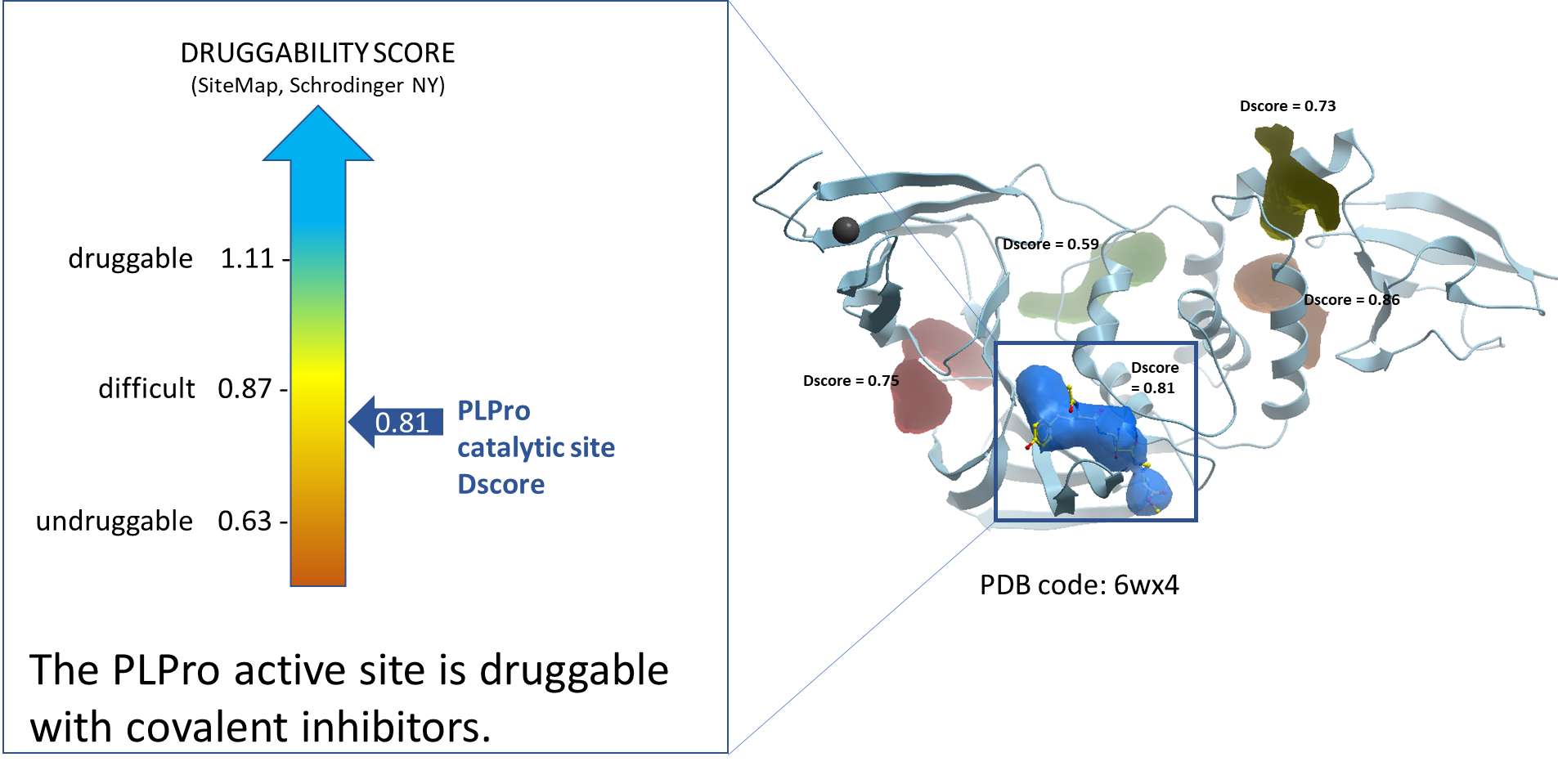
Figure 2. Druggability analysis of PLPro catalytic site. The blue pocket shows the catalytic site of the PLPro protein which is occupied by the non-natural amino acid-containing inhibitor, VIR251.
Using ICM, we found the amino acid sidechains facing towards the catalytic pocket and within its 2.8 Å vicinity. We identified twenty-four sidechains in total which include: W106, N109, N109, C111, Y112, L162, G163, D164, V165, R166, M208, S245, A246, P247, P248, Y264, N267, Y268, Q269, C270, G271, H272, Y273, T301, D302. Figure 3 shows these residues in sticks representation around the catalytic pocket of PLPro which is where VIR251 occupies.
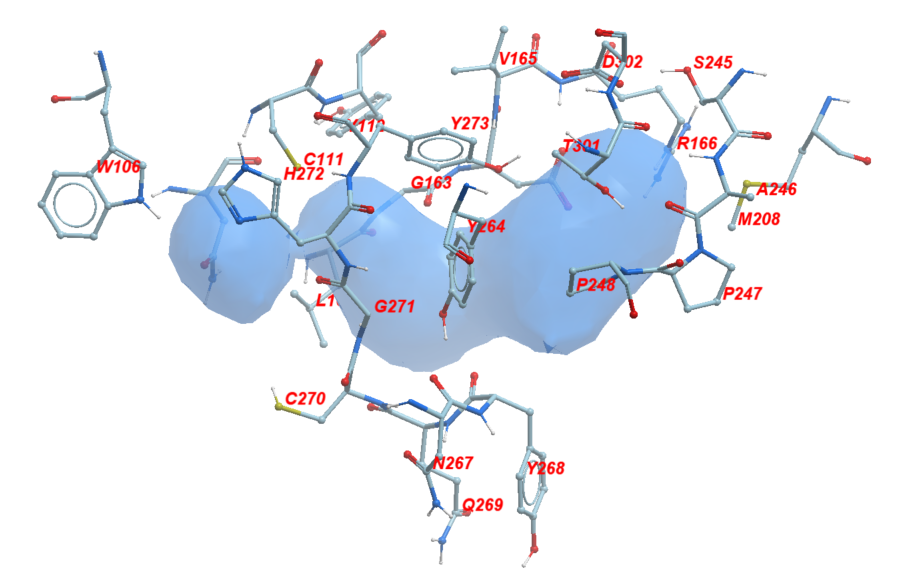
Figure 3. The neighboring residues lining the catalytic pocket of PLPro.
In my next post, I will show the sequence diversity across UniProt entries from the Alpha- and Betacoronavirus genera and map that to the PLPro’s catalytic site using its crystal structure.
As always, I would be happy to hear your comments. Please contact me via the “Leave a comment” link at the top of this post. Stay Tuned for my next post!
Reference:
- Rut, W. et al. Activity profiling and structures of inhibitor-bound SARS-CoV-2-PLpro protease provides a framework for anti-COVID-19 drug design. https://www.biorxiv.org/content/10.1101/2020.04.29.068890v2
