SARS-CoV-2 Helicase (NSP13) as a drug target:
The pandemic caused by SARS-CoV-2 is not over yet but instead has transformed into a chronic illness. So far, the expedited effort on drug development produced Mpro targeting drugs (Paxlovid) and repurposed RdRp inhibitor Molnupiravir. For effective long-term treatment, we intend to discover complementary antivirals targeting replication machinery such as viral helicase, in this case, NSP13. There are multiple precedences that helicase is druggable, starting with the discovery of HSV helicase inhibitor Pritelivir which showed an effective cure against acyclovir-resistant HSV (Birkmann, Bonsmann et al. 2022).
Virtual Screening
In parallel to the conventional medicinal chemistry approach (e.g., thiazoles and Brr2 series), we deployed virtual screening against the reported NSP13 structure (Newman et al., 2021). We practiced and established an iterative process to identify novel chemotypes with the optimal physicochemical profile. In this method, we docked 40 billion compounds from the Enamine REAL space and optimized the parameter with multiple filters (1a). End of these iterations (1b), we generated two sets of compounds commercially purchasable and de novo (generative AI) compounds. Later, we purchased 51 compounds from Enamine from the purchasable list and established the synthetic protocol for the de novo compounds (Scheme 1).
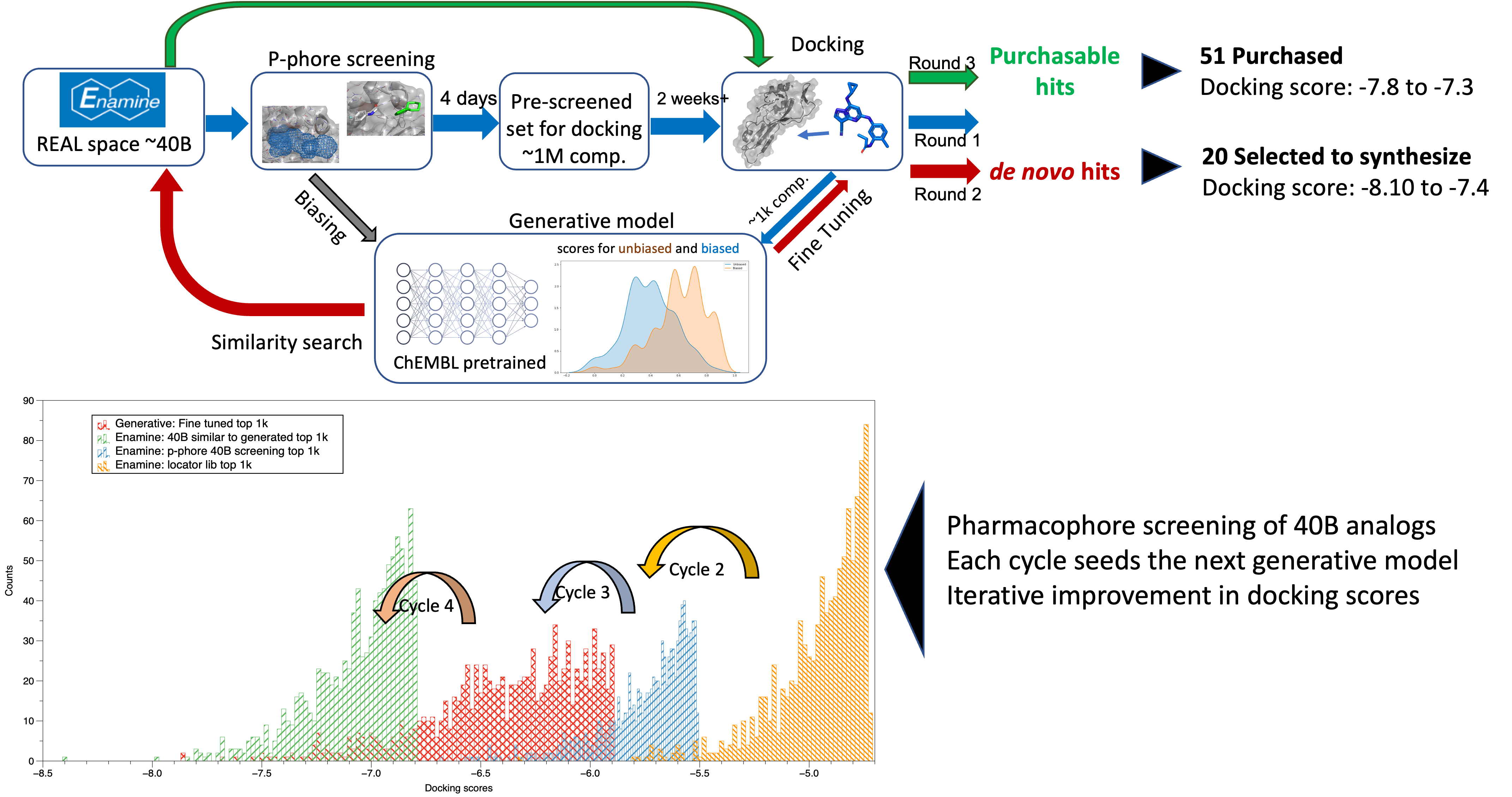
Figure 1: (top, a) Virtual screening workflow; (bottom, b) Iteration of VS with added filter for the better docking score
Purchasable N-Oxide Library:
Interestingly selected top 60 compounds contain a pyridine N-oxide functional group (we named as N-oxide series) connected with varying linkers and in a dipeptide structure (fig 2). Enamine successfully delivered 51 compounds from the requested 60 compounds (failed synthesis in the box).


Figure 2: List of purchasable compounds identified from docking against NSP13 (DS = Docking Score).
De-Novo Hits against NSP13:
Like the purchasable hits, the de-novo list also contains Pyridine N-Oxide among the majority of the top-scored compounds (e.g., fig 3). The de-novo (generative AI) compounds contain diamine, heterocycles, and aromatic diamine in between pyridine N-oxide and a para-fluoro pyridine acetic acid. The critical part of accessing these molecules is the synthesis of diamine linker. We are synthesizing in-house (with our CRO) in a multi-step synthesis (Scheme 1).
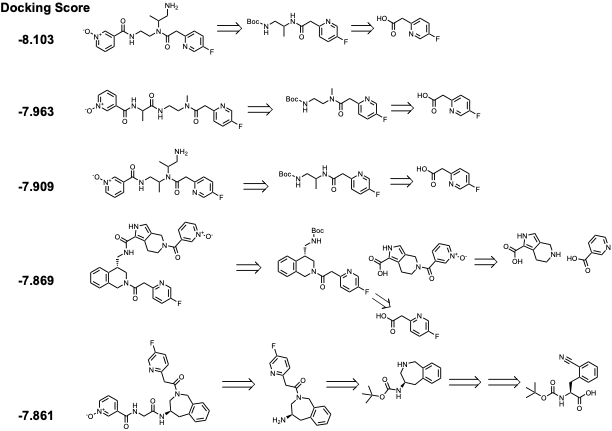
Figure 3. Retrosynthetic analysis of top-scored compounds from the de-novo hits (top five representatives)
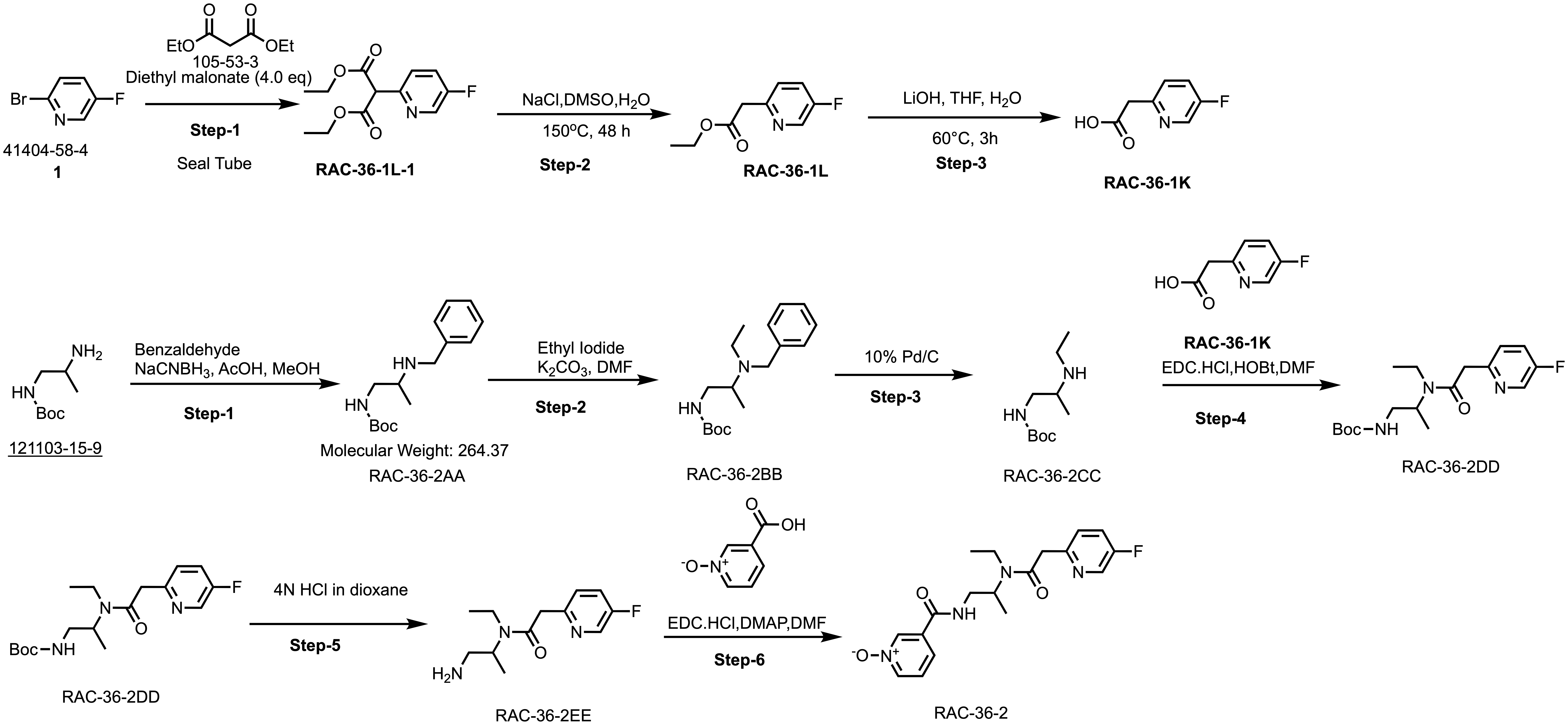
Scheme 1: Representative synthesis of de-novo compounds. [Chemistry Courtesy: CRO, Piramal Pharma Solution]
Helicase Hit Validation:
Upon purchase of the N-oxide series molecules, we screened them in multiple assays, including biophysical (SPR), enzymatic (ATPase), in-vitro antiviral assay, and X-ray crystallography. In the SPR assay, two N-oxides showed activity below 10 uM (Fig 4) against the SARS-CoV-2 and MERS Helicase. In the secondary ATPase assay, two compounds showed moderate inhibition below 50 µM. From this set of molecules, we further tested 6 compounds in a cell-based antiviral assay where none of the compounds showed any activity—further confirmation of the observed activity in SPR in the process (X-ray crystallography).
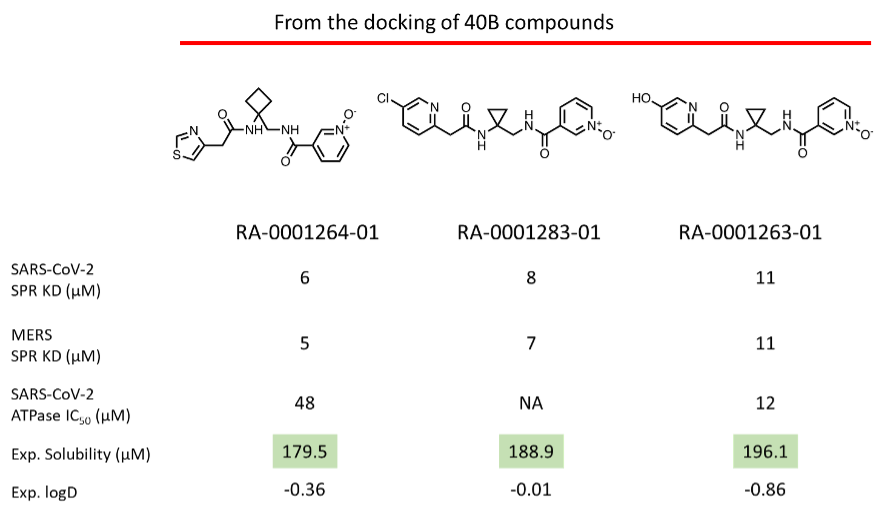
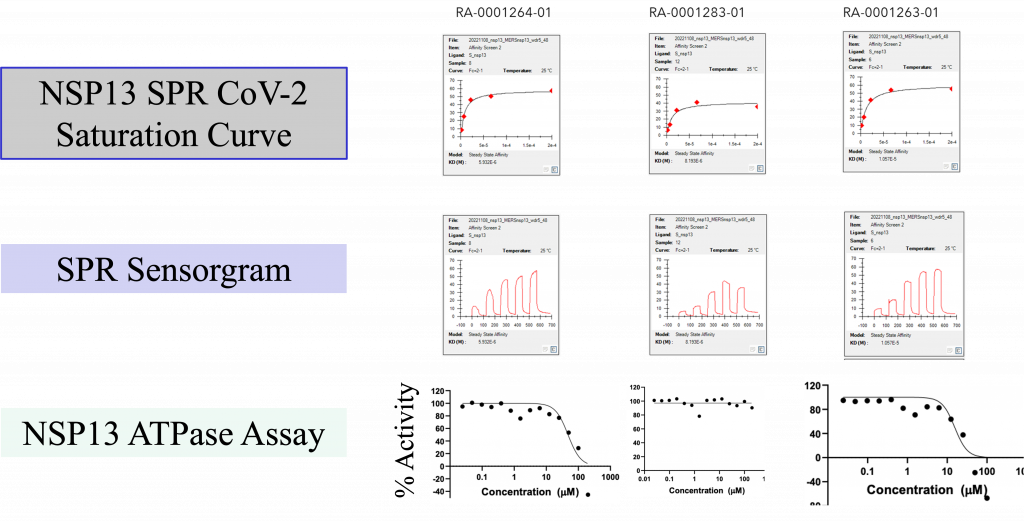
Figure 4: (Top) Top three N-oxide compounds with the activity profile; (Bottom) SPR sensorgram of the top three compounds against NSP13.
Strategies to confirm the activity of N-oxides and further advancements:
The N-oxides series molecules showed good physicochemical properties (clogP, cLogS, tPSA, logD7.4, solubility, etc.). But the observed activity is yet to confirm and validated. To do so, we initiated multiple approaches:
-
-
-
- Re-synthesis of the top hits (to improve the purity, as supplied materials are >90% pure)
- X-ray crystallography of the most active compounds in SPR (in the process)
- Deploy a small set of SAR
-
-
The activity of 51 compounds does not have a correlation between physicochemical properties and SPR activity. But it is indicative that a compound with optimal solubility and moderate cLogP (~1) has a better chance of being a potential hit.
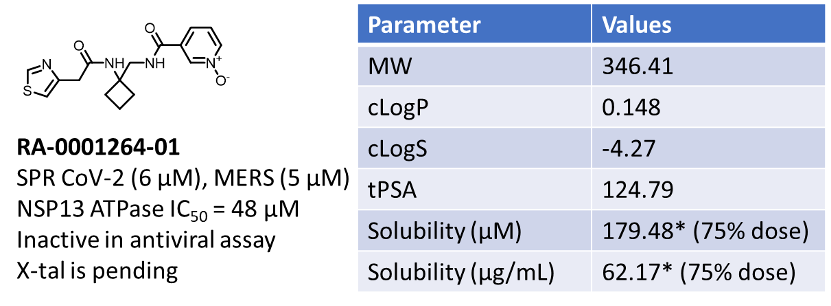
Figure 5: Physicochemical and activity profile of top hit
Rational of N-Oxide SAR design:
As the iterative docking showed, N-oxide is preferred for better interaction with the target protein; we want to test this hypothesis. We rationalize the following strategies:
-
-
-
- Replacement of N-oxide from the active scaffold (RA-0001264-01) with benzoic acid or nicotinic acid, which will confirm the necessity of the presence of N-oxide
- Replacement of N-oxide will make the synthesis much more tractable and speed up the library synthesis
- Alternate the N-oxide and thiazolyl acetic acid to confirm if a particular arrangement is preferred
- Replace the diamine linker with a simple or similar diamine
- Substitution of the N-oxide with other benzoic acid analogs
-
-
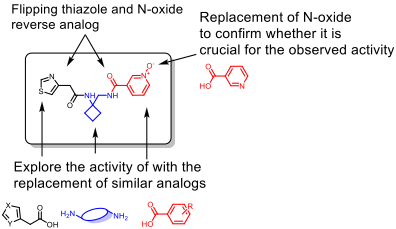
Figure 6: Proposed SAR map for the N-oxide series analogs
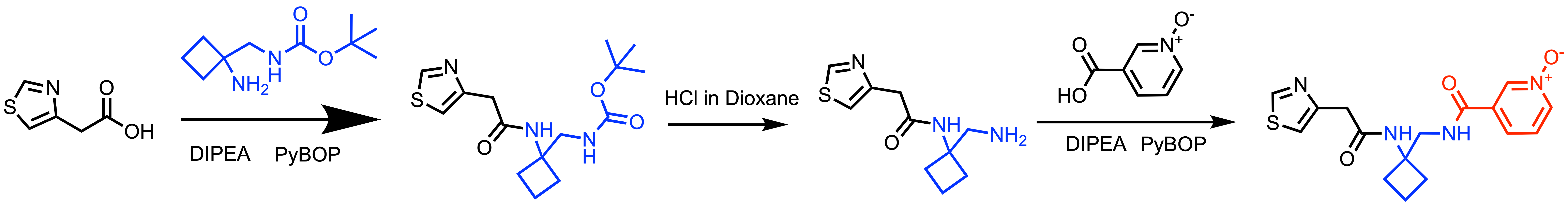
Scheme 2: General synthetic scheme for N-oxide library
With the planned SAR, we want to answer whether the predicted N-oxide is preferred for the observed NSP13 activity. Replacement of diamine with other amines guides us to explore more accessible diamine, and finally, thiazole modifications will confirm its importance as well as the mode of interactions.
Conclusions:
We started with a gigantic chemical space to identify a novel chemotype against NSP13. So far, we ended up with a few potential hits where the overserved activities need to be further validated through secondary assay. This hit confirmation will confirm the computational modeling, too!
**Viral helicase is a mystery to explore – stay tuned**
Acknowledgment:
- Virtual Docking: Konstantin Popov, UNC Chapel Hill, USA
- SPR and ATPase assay: Sumera Perveen, University of Toronto, Canada
- Compound Clustering and Selection: Mat’s Lab, Thomas Knight, UCL London, UK
- Chemistry Synthesis: Chemists @ UNC and Piramal Pharma Solution
To know more about READDI: https://www.readdi.org/
Interested in participating in AViDD Open Science? Check here

Anwar, congrats on this exciting work and on sharing your work-in-progress here. Could you maybe clarify which site of NSP13 was targeted in the virtual screening exercise?
Hi Matthieu,
Thank you very much. We did docking at the third site of NSP13, “C-terminus-B.”