As part of an ongoing collaboration with Meds4Kids Pharma (M4KPharma), we are testing a range of compounds identified and generated by them as being of interest in developing a molecule specific against ALK2 that has the right properties help treat some cases of DIPG. This also has significant implications for development of a drug that might help in FOP which also involves the dysregulation of ALK2.
Ros and I have been trying to crystallise some of the more interesting compounds from M4KPharma but we’ve run into a few problems. There are two constructs we use predominantly for crystallisation of new compounds as they have been shown to be robust and relatively well crystallisable. These constructs are of the kinase plus GS domain which we use in complex with FKBP12, and the kinase domain only which we use on its own. Both are expressed in insect cells and usually give a good robust expression that allows multiple compounds to be set up in crystal plates and significant chemical space explored from a single 2L purification. For the past couple of months our insect cells expressions have been failing to give us any expression of these two constructs which has made setting up crystal plates in high throughput impossible. Ros and I have been working with our Biotech team who are in charge of our baculo production facility to try and troubleshoot why this is the case; we’ve looked at whether the flasks have changed (they haven’t), the media has changed (it hasn’t), the virus has gone off and whether going back to an earlier virus stock might help (it didn’t). Currently I’ve got a pellet of the kinase+GS domain to look at, which was generated from a plate of revived constructs where the bacmids were recloned from very early in the pipeline. The cells were grown for 72h and harvested at 900g for 20 minutes. Hopefully that will have done the trick and we’ll have recovered the expression of our most useful ALK2 construct.
However, in the meantime these compounds still need investigating so I’ve been looking at crystallising three compounds (M4K2009, M4K3003 and M4K3007) with a construct of ALK2 which covers the kinase domain, the GS domain and contains the R206H mutation.
The purification went well and I managed to get enough from the prep to make up 12 plates – 4 plates for each of the compounds I’m looking at. Because the construct contains the GS domain, it is likely to require FKBP12 to stabilise it sufficiently to crystallise. Ros had some spare purified FKBP12 in the fridge which I could use for the complex. However, given the R206H mutation, the affinity for FKBP12 is decreased (but not eliminated – as you can see in panel A of figure 2 below) and means that the complex cannot be purified well by gel filtration.
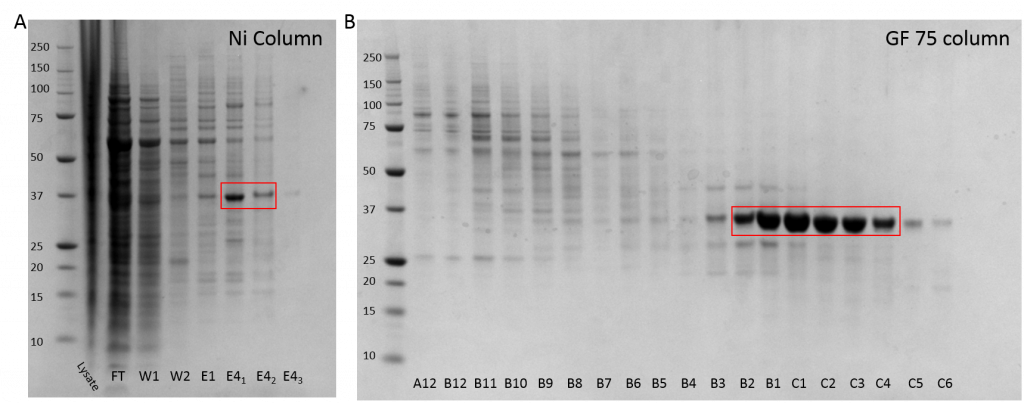
Fig. 1 A: Nickle purification of ALK2 (R206H) with a strong band observed in the first two Elution Buffer 4 fractions. B: GF75 purification shows the presence of protein in fractions B2 – C4.
Instead the proteins were mixed at a 1:1 ratio prior to having the compound added and set up in crystal trays. Four course screens were used (JCSG7, LFS6, HCS3, and HIN3) which sample a wide range of chemical space. JCSG7 and LFS6 were incubated at 20C while HCS3 and HIN3 were incubated at 4C.
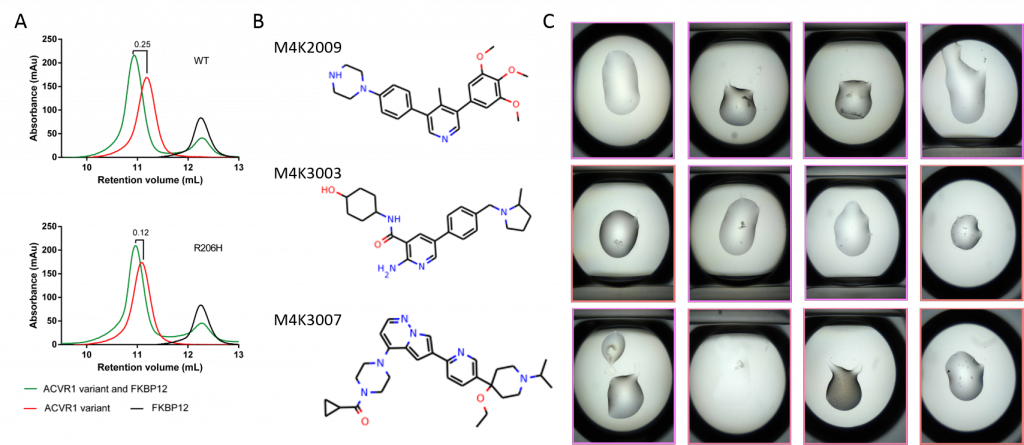
Fig. 2 A: Previous analytical gel filtration experiments show that the complexing of FKBP12 to ALK2 (wt) lead to a shift of 0.25ml suggesting a complex has been formed. When using the R206H construct, this shift is half of that of the wild type suggesting a decrease in affinity between the two proteins. B: The three compounds being investigated. C: Crystal plate drops showing potentially interesting crystalline matter for the ALK2/FKBP12 complex relating to the relevant compound from B
After 24h there are a couple of interesting drops that seem to have some crystalline matter in them which I will be watching with interest to see if they grow. I’m away next week so hopefully there will be something to see when I get back!
You can read more over on Zenodo.
