…and analysing their activity!
Following on from the work I talked about here I’ve been able to start looking at the influence the different phosphorylation sites have over the ability of ALK2 to activate SMAD1.
Just coming through the SGC expression pipeline are several constructs containing mutations in various combinations of the sites known to be phosphorylated in and around the ‘GS loop’ of ALK2. This will prevent phosphorylation on that site and the change that makes to the activity of ALK2 will tell us something about how that site influences ALK2 function. These are shown in the figure below. Here they’re all labelled as ACVR1 due to our internal system using the gene name rather than the protein name.
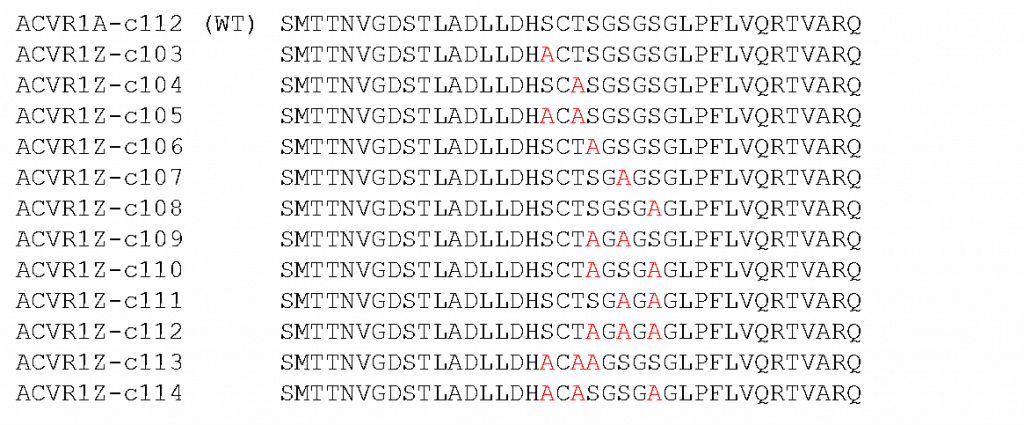
A list of ALK2 constructs containing mutations in the GS loop region of the protein. The mutations are from either Ser or Thr to Ala to knock out phosphorylation on that site.
One of the key findings from my previous experiments was that there seemed to be some sites that needed to be phosphorylated for maximal activation of SMAD – for example when a kinase dead type II binding partner was used some sites were not phosphorylated on ALK2 and this also corresponded to a reduction in SMAD1 phosphorylation. These mutants should help me explore which sites are vital for ALK2 activity and ultimately help me understand why the FOP mutation causes increased activity.
Anyway… now that these have been cloned into a baculo expression vector I can start actually purifying the protein and doing some experiments with them.
I had 1l of c103 cells and c104 cells which I purified with fairly high yields. I also had 1l of c105 cells but something went really wrong with the purification and I ended up not having enough to do any actual experiments with. I arranged to get some more c105 cells and 1l c106 cells but while those were being grown I went ahead and did a small scale experiment looking at how c103 and c104 behaved in comparison to the wild type ALK2.
I used my standard assay for monitoring SMAD1 phosphorylation. I incubated SMAD1 with ALK2 and with a type II receptor (in this case ACVR2 and BMPR2) at high enough concentrations to allow receptor association under normal circumstances. I then added activating quantities of ATP along with DTT, Mg2+ and Mn2+ ions necessary for activity and took 1ul samples at regular time intervals up to 30 minutes. The samples were quenched in MS buffer (which contains enough acid to stop any phosphorylation by a kinase) and at the end the results were run on the mass spec machine. Full details of the experimental protocol and the purification can be found over on Zenodo.
The results were fairly interesting but without other experiments remain somewhat inconclusive.
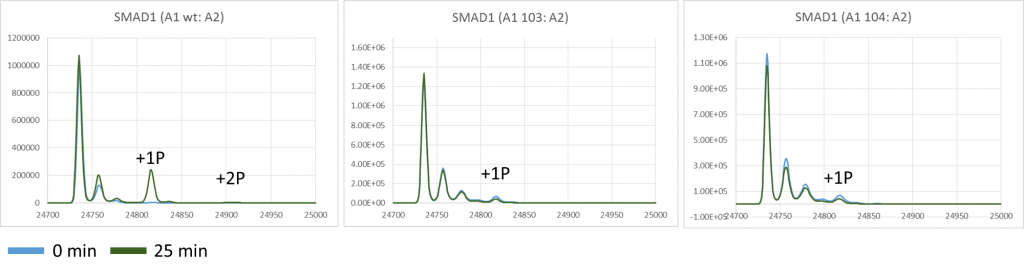
Graphs showing the mass peaks detected by mass spec for SMAD1 in its unphosphorylated state and in various states of phosphorylation at +80 Da intervals at both 0 min and 25min for ALK2 wild type, ALK2 c103 and ALK2 c104 mutants.
What I can see is that compared to wildtype ALK2, it looks like there is much reduced phosphorylation on SMAD1 using the phosphomutants over 25 minutes.
I also looked at the phosphorylation of ALK2 in this experiment which can be seen in the figure below. This shows that there is a reduction of one occupied phosphorylation site compared to WT which is to be expected given one site has been removed by mutation however given we know from other experiments that there are up to 8 possible phosphorylation sites it’s not clear if the mutation is influencing which of these sites then is occupied.
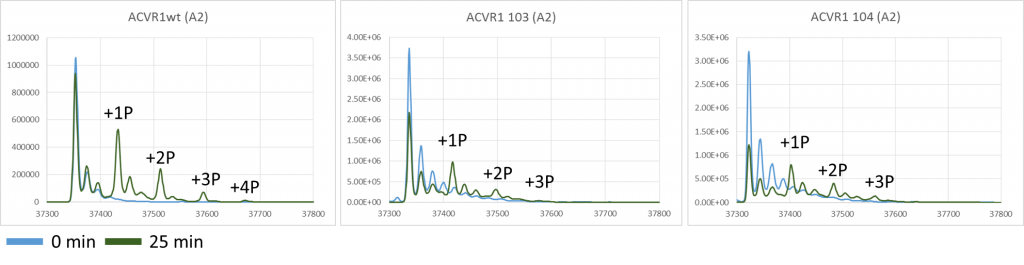
Graphs showing the mass peaks detected by mass spec for ALK2 in it’s unphosphorylated state and in various states of phosphorylation at +80 Da intervals at both 0min and 25min for ALK2 wild type, ALK2 c103 and ALK2 c104 mutants.
There are questions unanswered however, such as do the mutations reduce the affinity between the type I and type II proteins such that a higher concentration might be needed to induce association (given that we already need to artificially induce their association via high concentrations to make up for the lack of membrane co-localisation that would occur in the cell) and if left for longer, would they show some activity rather than practically nothing? This suggests my next line of experimental inquiry which will be to alter the concentration of proteins in my experimental setup to see if increasing the concentrations of ALK2 and ACVR2 make any difference to the results in comparison to a wild type control. I’ll also let the experiment run for longer to see if this gives any indication as to whether the activity has been stopped or merely reduced. Increasing the time will also help pick up any differences in ALK2 phosphorylation as it may allow more sites to become occupied and thus pick up any limitations that the mutation is putting on that aside from the removal of one site by the mutation its self.
I’ll also think about looking at SMAD1 phosphorylation by western blot in addition to mass spec as we have a good antibody for looking at specifically phosphorylated SMAD1 which will show up very small amounts of phosphorylated SMAD1 that the mass spec might miss.
For details on the experiments you can find it over on Zenodo here:
