I used a surface plasmon resonance assay to determine the binding affinities of hit compounds that were identified in 19F NMR spectroscopy screens against the zinc finger ubiquitin binding domain of USP5. To see how these assays work, check out some of my previous posts! You can see details of the SPR experiment on Zenodo.
Compound UBXML78 had the best binding affinity, with a KD of approximately 59 µM and a ligand efficiency LE=0.24 (Figure 1). DAT76 (KD ̴ 109 µM) had the best ligand efficiency (LR=0.29) (Figure 2). These compounds are rather weak but together with my previous crystal structures, still provide a solid starting point for a hit optimization campaign.
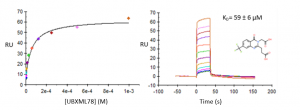
Figure 1. Representative binding curve of UBXML78 and USP5 Zf-UBD
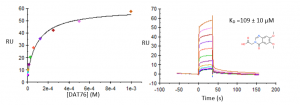
Figure 2. Representative binding curve of DAT76 and USP5 Zf-UBD
Now that I’ve tested all the hit compounds from the 19F NMR screens using SPR, I wanted to see if I could correlate the 19F NMR assay results with the SPR binding affinity measurements. I find that a chemical shift greater than 1 ppm in the 19F NMR assay results in binding affinities less than 400 µM. I can use this cut-off to select compounds for SPR based on the NMR screen. This will limit testing of very weak compounds by SPR.
Next, I’ll be doing hit expansions of a few of the compounds of interest where I will run substructure searches and docking simulations to select commercially available compounds for the next set of screening. This will hopefully increase my understanding of the structure activity relationship (SAR) of the chemical series, which in turn will inform the rational design of more potent USP5 Zf-UBD ligands. Stay tuned!
