Structure Solution for BMPR1B/FKBP12 complex bound to M4K2009.
Good news everybody!
From the crystals I sent to the Diamond Light Source (a synchrotron, or particle accelerator, that gives very high quality x-rays) last week, I got a data set for BMPR1B/FKBP12 with M4K2009 bound to it! This is one of the compounds that the M4K Pharma company is very keen to see in situ to understand it’s action. While it would ideally be bound to ALK2 (something Ros is working on), seeing it bound to BMPR1B is also useful for understanding it’s selectivity (or lack of it) across various BMP signalling proteins.
The data looks like it refines down to around 2.34 Å. I’ve then spent much of last week analysing the data and trying to get a decent model of the proteins.
The data is in the P1 21 1 crystal form (this is just a way of describing the symmetry within the crystal) and has two copies of the complex in the asymmetric unit (which is another way of defining how the protein is packed inside the crystal). It’s refining nicely and I’ve got a very nice clear look at the compound bound to the binding site however I also noticed that several (but not all) exposed cysteines have been covalently modified by something in the plate well condition to give very distinct blobs of electron density that need to be identified.
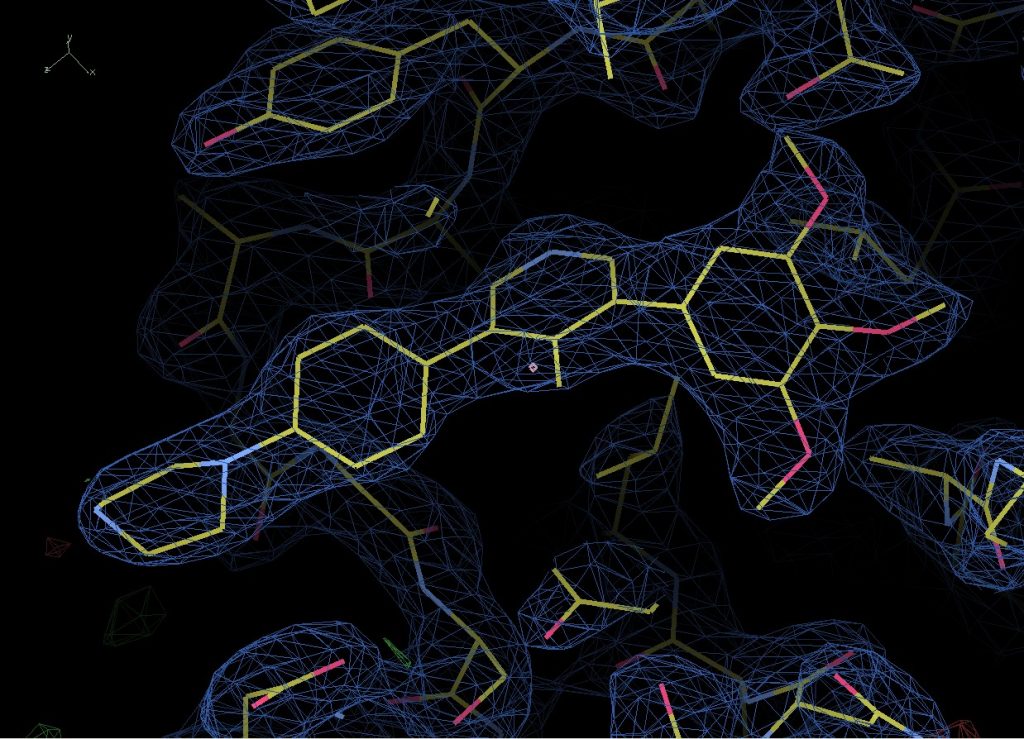
Compound M4K2009 bound to BMPR1B.
Looking back, the crystal was grown in 20% PEG8000, 0.2M Magnesium Acetate and 0.1M Cacodylate pH6.5. Under these conditions it’s most likely that the cacodylate has reacted with the cys residue, most likely catalysed by neighbouring side chains deprotonating the SH group and allowing it to react with the arsenic at the centre of Cacodylate. However, there are two possible reactions that could occur, one of which leaves you with a sulphur bound to an arsenic with two methyl groups and an oxygen attached to it, and the other which leaves you with just two methyl groups bound to the arsenic. Now – consulting with my chemist colleagues they believe that the oxygen containing result is most likely and I’m inclined to agree as it is the simplest and most easily catalysed reaction. However, looking at the density and modelling in both options, it looks like the second might fit better. Either there is a more complex reaction occurring or it’s possible the oxygen has been knocked off by radiation damage during data collection.
Currently I’ve modeled them all as the second option since as a general rule what the data is telling you should win out over the prediction, but I can relatively easily change that if necessary. I need to get a second opinion on it at some point and any thoughts are greatly received.
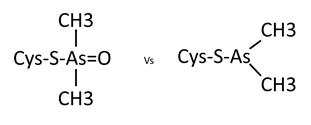
Two possible options of cacodylate modification to cys residues.
Fortunately, none of the cys residues are near the binding site however so this isn’t an issue that is going to effect that result and I can be fairly confident that compound binding is not influenced by it.
You can read more details on the processing on Zenodo.
