A belated Happy New Year everyone! It has been a good while since my last post so I wanted to update you on what I have been spending 2019 on so far.
After identifying some really promising follow-up compounds in my activity assay, the next step was to find out how they bind to HAO1, as illustrated in figure 1. Specifically I want to know how different chemical constituents of the new compounds interact with and/or cause conformational changes to amino acids within the HAO1 structure. By understanding which groups improve binding (e.g. that stabilise the bound state by interacting with nearby amino acids), which group increase inhibition (e.g. by disrupting active site residues like Trp110) and which groups are not required for the compound to work (i.e. no apparent interaction with the protein), we can rationally design compounds that combine the most effective chemical groups to make more potent inhibitors.
In practical terms, this means that I have made >250 HAO1 crystals and soaked each of my follow up compounds twice (127 compounds times 2 crystals each = 254 crystals soaked). This duplication helps ensure complete coverage of all compounds (e.g. if one replicate is not mounted/collected correctly) and also improves confidence in my results i.e. if I see the same compound making the same interactions in two independent structures, I feel confident that the interactions I see are significant changes dependent on compound-binding.
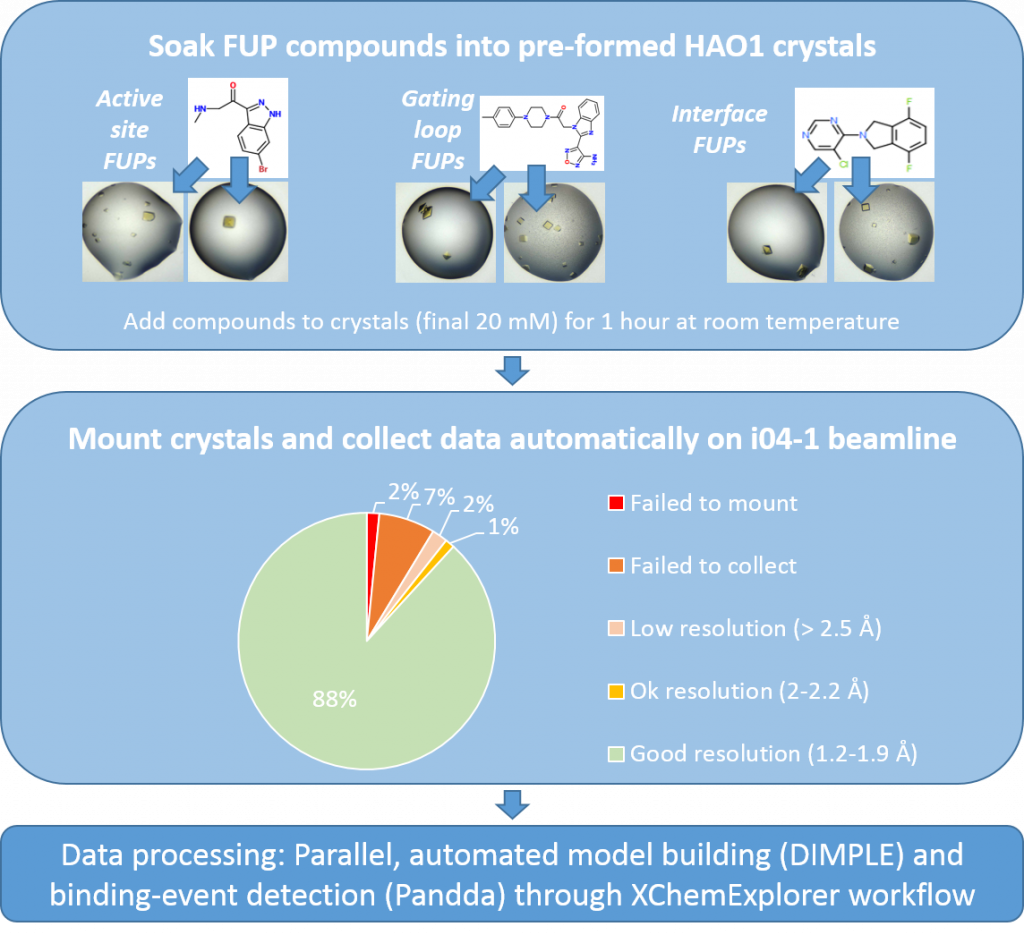
Figure 1: Experimental workflow for characterising follow-up compounds by x-ray crystallography. Follow-up compounds from fragments 1, 2 and 5 are described here, whereas those from fragment 6 are described here. Completed experiments are in light blue boxes and ongoing experiment is in dark blue.
I have collected a total of 227 good datasets (resolution range of 1.2 to 2.2 Å, 90% better than 1.7 Å) and now I am hard at work processing these through the XChemExplorer pipeline, ready to tell you all about the compound-bound structures in the next few weeks.
