Over the past few weeks I’ve been perfecting a method for screening a small set of therapeutic compounds on DIPG cell lines. Before starting I made sure to discuss the techniques used in other groups to make sure that I design an assay that is comparable to theirs, although I may make my own changes later, and see if this makes a difference to how effective drugs are.
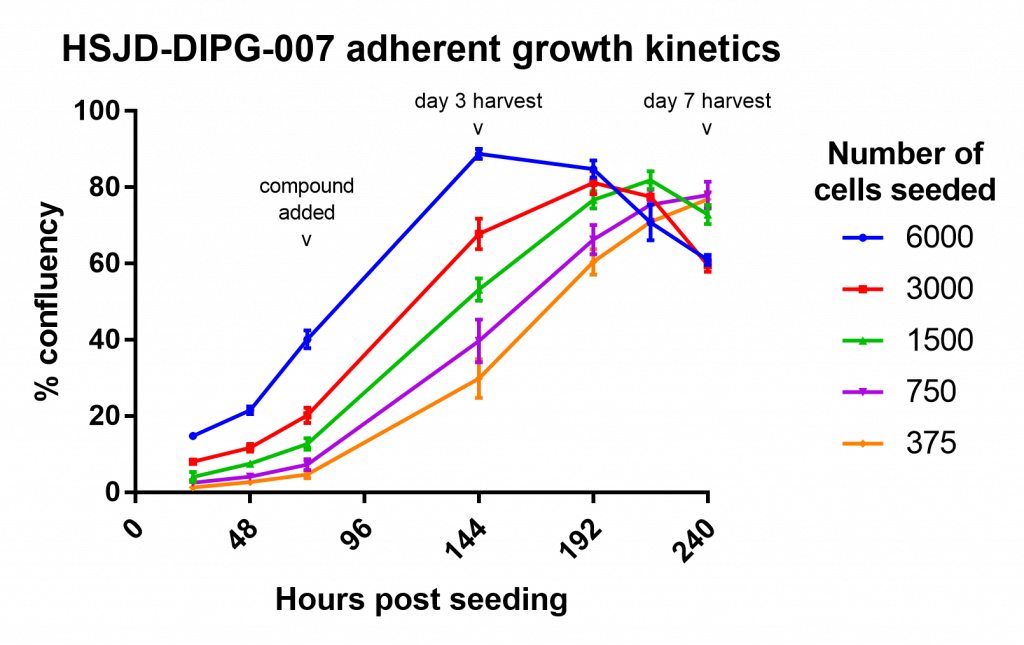
My first attempt wasn’t useful as a drug assay, but did allow me to measure the growth of the cells over time:
This was a great help for making choices about how many cells to seed and what time periods to use in the assay.
Eight iterations later, I now have an initial proof-of-concept dataset, and a (mostly) final protocol. The general scheme of the assay is as follows:
Coat the plates with laminin, a protein that allows the cells to stick to the plate, instead of growing as floating spheres (which is the current growth mode of choice in the field). Seed 2,000 cells into each well, in their usual growth medium, and let them grow undisturbed for 3 days. I then add a range of concentrations of each drug and a marker for cell death to the wells (with three repeats each). After four days, I add a solution (called CellTiter-Glo) that contains a luciferase enzyme and therefore glows in the presence of ATP. As ATP is only found in live cells, this light provides an indication of the number of live cells present.
Once I detect the luciferase glow with a plate reader, I can plot a curve of how many cells are alive at the end of each treatment (or, how many are dead if I look at the cell death marker), which looks like this:
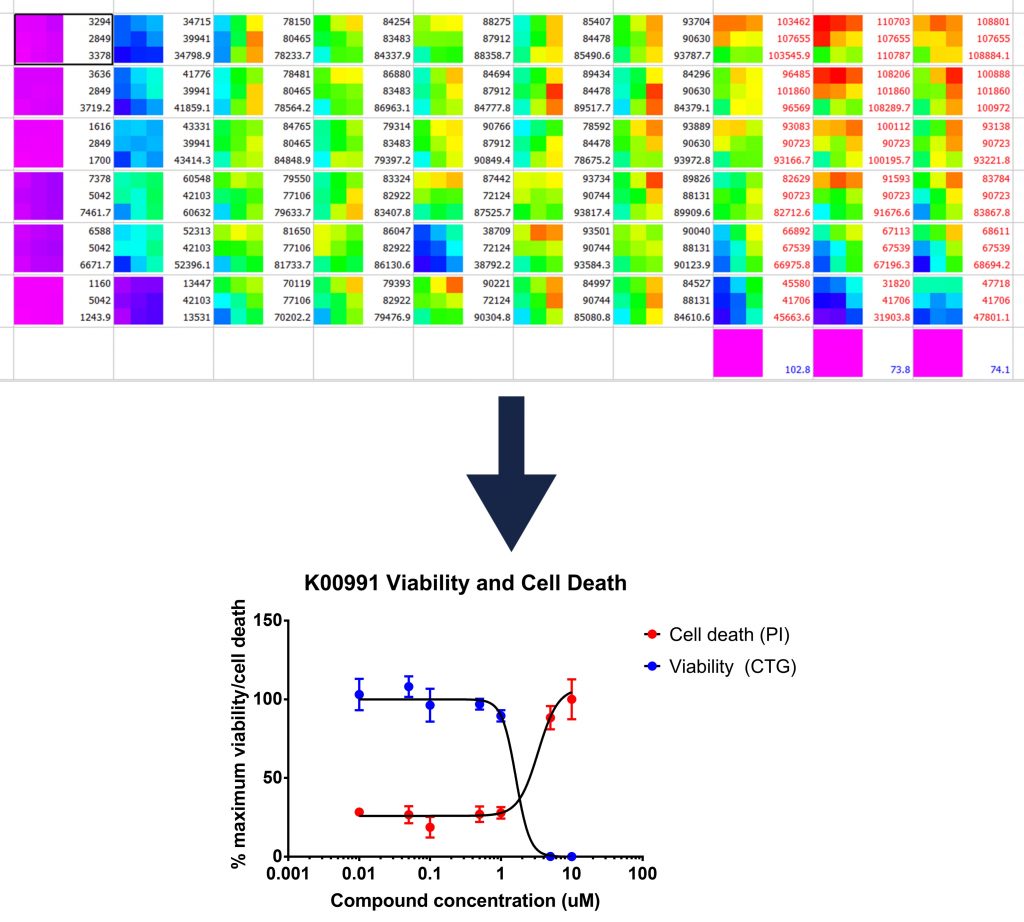
Top: a visual representation of the luminescence readings recorded
Bottom: a graph produced from such readings which illustrated the reduction of cell viability caused by drug treatment
For this compound, the assay worked well, however with another I noticed that I only seemed to be able to see inhibition for three days, and not four. This could be because the drug degrades or is digested over time and therefore that I may need to add the drug more than once in future .
You can read the complete method, and the full dataset on this page.
P.S. If anyone has tips on how to present each biological repeat, my ears are open !

Hi Liz!
I was wondering, since your readout is Celltiterglo anyway, why do you choose to do the assay on adherent cells as opposed to the original spheres? CTG works very well on spheres, and there are several reports showing that adherent cells do not respond to compounds with the same sensitivity as spheres. I work on spheres myself so I’m always glad to hear perspectives/comments from other sphere users 🙂
Cheers
Evelyne (SGC Toronto)
Hi 🙂
I’m initially doing two dimensional assays so that I can compare my results to those of other groups, because this is the standard they use (and have published), but I’ll also be doing 3D assays to compare the results (which is what I was optimising in this post: https://opennotebook.thesgc.org/making-single-dipg-spheres/ )
Do you do all your assays in three dimensions?
Yes I do all my assays in spheres or organoids, and sometimes when I have a hit, I try it in the same patient line but grown in 2D and the response to some of our inhibitors is very different (adherent can be more resistant to some probes). 3D models are not as easy to work with but more relevant I think! I don’t have that many assays for them though, mostly viability (CTG or flow cytometry), limiting dilution assays. I’d love to do more imaging! I think it’s really cool that you tried PI in your spheres, I was actually going to try the new cytoplasmic incucyte dye at some point, they claim they’re non toxic 🙂