Hi Everyone,
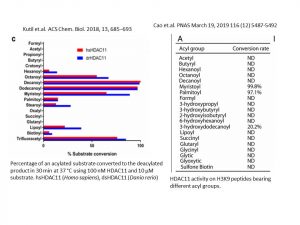
This is the first time I am going to share with you my research and I will start with the project I am currently working on, which is the development of cellular HDAC11 target engagement assay. HDAC11, the only member of class IV HDAC subfamily, was believed to be responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). However, recent in vitro data indicates that HDAC11 is 10,000 times more efficient in lysine defatty-acylation than deacetylation (Fig1).
Fig1.
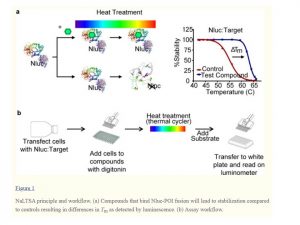
Target engagement assays allow us to test if potential inhibitors enter the cell and interact with a target protein. Here, we are interested in developing this type of assay for HDAC11. Recently, novel potent and cell active HDAC11 inhibitors have been developed, which can be used for cellular assay validation (FT895 and compound 14). (PMID: 29776742). The activity of inhibitors in cells were tested using NanoBRET™ Target Engagement Assay, which measured the apparent affinity of test compounds by competitive displacement of proprietary compound labeled with a fluorescent tracer reversibly bound to HDAC11 fused to Nanoluc luciferase. Currently we are in a process of development of our own fluorescent tracer based on published HDAC11 inhibitors and hopefully we will be able to use it for HDAC11 cellular assay development. However, since the tracer is not currently available I had to try other methods. One generic method for confirming target engagement is Cellular Thermal Stability Assays (CETSA) in which binding of the inhibitor increases the thermal stability of the target protein. I tested thermal stability of HDAC11 upon binding to published HDAC11 inhibitors using NanoLuc technology (PMID: 29937980). In the assay you are transfecting cells with protein fused to Nanloluc luciferease, treat with inhibitors and measure bioluminescent signal after heat denaturation. Compound binding to the protein should lead to protein stabilization compared to controls. The system can be used in cells, which are not permeabilized and permeabilized with digitonin as a control (Fig2).
Fig2.
ACS Med Chem Lett. 2018 Jun 14; 9(6): 546–551
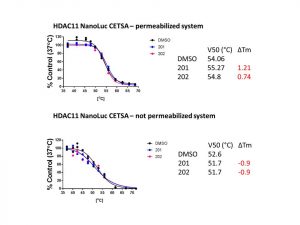
In this experiment, I did not observe thermal stabilization of HDAC11-NanoLuc upon compounds FT895 (202) and 14 (201) treatment (Fig3). Experimental assay details are posted on Zenodo.
Fig3.
Next I am going to test if I can detect any changes in histones acetylation upon HDAC11 overexpression and inhibitors treatment. I will also update you with my effort to develop assay based on two findings:
- HDAC11 suppresses the thermogenic program of adipose tissue (PMID: 30089714).
- HDAC11 regulates the expression of intereukin 10 (PMID: 19011628).

Dear Magdalena
you probably have recombinant purified protein of hdac11 available. did you test the tool cpds used in CETSA (i assume that is the 201 and 202 curves) on purified protein. just to be sure that there is not something else wrong with the cpds
best alex
The compounds work in our in vitro assay (IC50 in nM range), but I do not have any proof that they really work in cells. In original paper the authors mentioned that cellular IC50 is lower than 20 nM based on results from Nanoluc tracer experiment, but I have not seen any data on this.
Recent paper (https://pubs.acs.org/doi/pdf/10.1021/acschembio.9b00292?rand=jqrhoi7j) showed that FT895 is not affecting SHMT2 palmitolylation up to 50 uM. I would like to try sis17, the novel HDAC11 inhibitor in this system and I am currently taking totally different approach for assay development. Soon I should be able to post the data.
Calculate the Delta AUC and plot the Delta AUC to the R2 value. This is a better measurement of the shift. Also with the Nano CETSA the shifts are very small compared to traditional CETSA.
Thank you. I will try. I currently stopped using NL-tagged constructs. CETSA with HiBIT/LgBiT system gives my much better results.
Really interesting method. Could this difference in assay sensitivity be attributed to the higher intrinsic stability of nanoluciferase compared to the split nanoluciferase in HiBIT/LgBiT system?