The experimental set-up and results covered in this post can be found here: 10.5281/zenodo.1218375
The etiology of HD is the expansion of Q repeats or CAG repeats in the HTT gene, which produces the huntingtin protein. This causes improper folding of the protein and leads to protein aggregation in neuronal cells, which ultimately causes the movement, cognitive and psychiatric impairment we see in people with HD. You can read more about HTT and HD in my earlier blog post here (https://openlabnotebooks.org/background-on-htt-and-hd/).
Although we know that HD is caused by mutant HTT, no one has yet developed a cure for it. As I am investigating the effects the number of Q repeats in HTT has on the biological function in cells, I will attempt to recapitulate the disease in a cell model by introducing exogenous HTT with different numbers of Q repeats into mammalian cells and studying the phenotype of the cells.
In my previous post, I managed to use baculovirus to overexpress wild-type HTT tagged with FLAG in mammalian cells (read more about my optimisation of baculovirus transduction here: https://openlabnotebooks.org/overexpressing-htt-in-mammalian-cells/). Using the same pipeline and protocol, I tried expressing HTT with different lengths of Q repeats in mammalian cells.
D1: 23 Q repeats
D2: 54 Q repeats
D4: 79 Q repeats
D6: 145 Q repeats
Generally, a normal person would have up to about 35 Q repeats in HTT while people with HD have more 45 Q repeats, with the severity of the disease increasing with the number of Q repeats. Here is a set of graphs I used in my earlier blog post on the background of HTT and HD which you can use to correlate severity of the disease (measured by the age of onset and age at death) with the number of Q repeats in the constructs I am using for my experiment.

Figure 1, Keum et al., Am J Hum Genet 2016
As with all the other baculoviruses used in my experiments, these HTT baculoviruses were developed here at SGC Toronto by Rachel Harding, Peter Loppnau and Alma Seitova, who designed, cloned and produced the HTT baculoviruses.
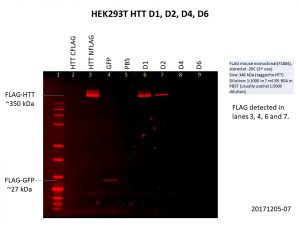
As you can see, I included the HTT C-terminal FLAG-tag and N-terminal FLAG-tag baculoviruses used previously as controls and I got the same results as I did previously, only the N-terminal FLAG-tag was detected. I also included the GFP control.
The D1 construct was expressed quite well, but D2 was not as good, and there was almost no expression of D4 and D6. This is not so surprising as the constructs are much bigger with increasing number of Q repeats, making it harder for both the delivery of the construct into the cells and the expression of the construct once it gets into the cell. Here’s another image showing the levels of HTT in the cells, with GAPDH included as the loading control.
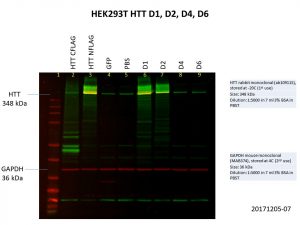
As with the previous image showing FLAG expression, HTT expression in D4 and D6 are comparable to that of the GFP and PBS controls. More optimisation will be needed to further improve the expression of these larger constructs.

Jolene, if pathogenic variants of Huntingtin are misfolded, should their baculo-expressed constructs be soluble in mammalian cell lines?
Great question! Actually, that might be one of the possible reasons we get such low expression levels with the larger constructs. It might be possible to try detecting aggregates in the cells.
Jolene Ho, Are eye-motifs epigenetic switches? (NATURE Chemistry) MSN Short video clip seen here) https://www.msn.com/en-us/video/wonder/new-dna-structure-discovered-inside-living-human-cells/vi-AAwfiyx?ocid=spartanntp If so, where’s the eye-motif for HTT? Enjoy 🙂
Hey J.B. Shaw, thanks for the video, that’s really interesting! I suppose the paper the video is referencing is this: https://www.nature.com/articles/s41557-018-0046-3? I personally didn’t know anything about i-motifs until I watched your link and did some research. It seems like the i-motif was discovered quite some time ago, in 1993, although it has not be shown in living cells until now. I believe this is the article in which i-motifs were first described, but please correct me if I am wrong: https://www.nature.com/articles/363561a0.
Anyway, the video you linked mentioned that the i-motifs were observed to appear and disappear, and that the i-motifs might be involved in turning genes on and off. This would strongly suggest that the i-motifs are acting as epigenetic switches.
As for the links between HTT and the i-motifs, I don’t think anyone has looked at that yet (again, please correct me if I am wrong). So far, people have attributed the etiology of HD to the misfolded mutant HTT protein. However, the mutant HTT is misfolded due to the expanded CAG region, and the i-motifs occur in cytosine-rich regions. I’m not sure if the expanded CAG region of mutant HTT is cytosine-rich enough for i-motifs to form, but it would be very interesting if such motifs were found in the mutant HTT gene!