Hi all, it’s been a while since my last post; I’ve been busy in the lab purifying new protein constructs, developing a new assay and expanding the chemical series of my weak binding hits for USP5 ZnF-UBD, so there will be a couple of posts coming your way in the following weeks.
This week, my post focuses on my efforts to expand the chemical series against USP5 ZnF-UBD to continue to understand the structure activity relationship (SAR) and improve potency. I used GLIDE to dock commercial analogues of hit compounds DAT00000194a and DAT00000201a (Table 1) as well as fragments from my selection of ligands from my work at Molecular Forecaster (MFI). The source library for the search for analogues of DAT00000194 and DAT00000201 was Enamine’s REAL database of 700 million chemically accessible compounds. You can see details of this work on Zenodo.
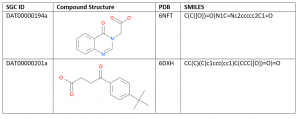
Table 1. USP5 ZnF-UBD Hits
20 commercial compounds were selected and ordered based on docking pose, protein-ligand interactions, docking score, chemical groups, and cost. For example, an analogue of DAT00000201a was chosen based on the hydrophobic and hydrogen bond interactions of the chemical group extending from the phenyl ring. The docked pose suggests a potential hydrogen bond interaction with residue D264 and hydrophobic interactions with residue M266 (Figure 1).
You can see some of the selected compounds at the end of this post, and download the sdf file!
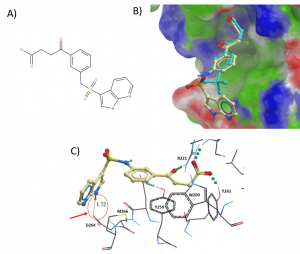
Figure 1. a) Analogue of DAT00000201a b) Surface representation of USP5 ZnF-UBD with DAT00000201a (cyan) and docked pose of analogue compound (tan) [PDB: 6DXH]; c) Ribbon representation of USP5 ZnF-UBD and docked pose of analogue compound [PDB: 6DXH]
It’ll be interesting to see how well the analogues bind to USP5 ZnF-UBD, and how the crystallized pose of the ligands will compare to the docked pose. When the compounds arrive in a couple of weeks, I’ll be testing these compounds using a surface plasmon resonance assay. Stay tuned!
| Structure Image | Smiles | Mol. Weight |
|---|---|---|
| O=C([O-])CCC(=O)c1cccc(NS(=O)(=O)c2c[nH]c3ncccc23)c1 | 372.4 | |
| Cc1ccc(C(=O)[O-])cc1S(=O)(=O)Nc1cccc(C(=O)CCC(=O)[O-])c1 | 389.4 | |
| C[C@@H](CC(=O)c1cccc(-c2ccc3c(c2)CCCO3)c1)C(=O)[O-] | 323.4 | |
| CN(c1ccc(S(C)(=O)=O)cc1)S(=O)(=O)c1ccc(C(=O)CCC(=O)[O-])cc1 | 424.5 | |
| O=C([O-])CCC(=O)c1cccc(NS(=O)(=O)c2c[nH]c(C(=O)N3CCCC3)c2)c1 | 418.5 |
