In my last post, I developed a low-throughput 19F NMR screening assay against USP5 Zf-UBD, where ligand binding of a ubiquitin peptide was detected by perturbations in the resonance of a fluorinated tryptophan at the binding pocket. I recently screened the first batch of compounds that Ivan Franzoni and Renato Freitas, post-docs at the University of Toronto and SGC, respectively, selected by virtual screening. You can find details of the NMR screening results on Zenodo.
As mentioned in my last post, USP5 Zf-UBD was fluorinated at two positions, W209 and W197. W209 is involved in coordinating ubiquitin in the binding pocket of USP5 Zf-UBD, whereas W197 is remote from the binding pocket (Figure 1). In the fluorinated USP5 Zf-UBD, two peaks of equal intensity are measured at approximately 118 ppm and 225 ppm. In the presence of some small molecules, only one of the two peaks, at 118 ppm, is shifted to the right with a decrease in peak area. Peak 2 at 225 ppm remains unchanged in intensity and chemical shift (Figure 2).
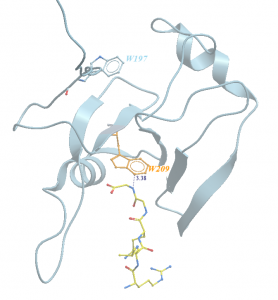
Figure 1. Ribbon representation of tryptophan residues of USP5 Zf-UBD (grey) and ubiquitin (yellow)
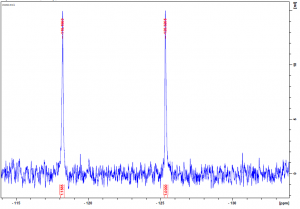
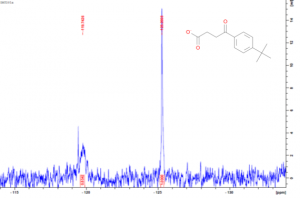
Figure 2. 19F NMR spectra of a) USP5171-290 b) 1:25 USP5171-290 :DAT201
In general, the compounds that resulted in the largest chemical shift to the right from the control fluorine resonance also had the greatest decrease in peak area relative to peak 2. Of the 33 compounds screened, 12 of the compounds showed significant changes in peak 1 fluorine resonance. The purity of these compounds of interest was >95% as measured by UPLC-MS. The high “hit” rate is likely due to the high compound concentration, the sensitivity of the 19F NMR assay and the biased pool of compounds that were predicted to be favorable for USP5 binding. Overall, 19F NMR gives insight into the structural perturbations of the USP5 binding pocket when small molecules bind. Next, I will have to confirm binding in an orthogonal assay and use a more quantitative method to determine binding affinities, such as a surface plasmon resonance assay (SPR). Once I have a better understanding of the Kd of these compounds, I can prioritize the best binding compounds for crystallization studies, from which I can then guide molecular designs of the ligands to increase binding affinity.

4 Replies to “Compounds of interest identified by screening a focused library against USP5 Zf-UBD with a 19F NMR assay”